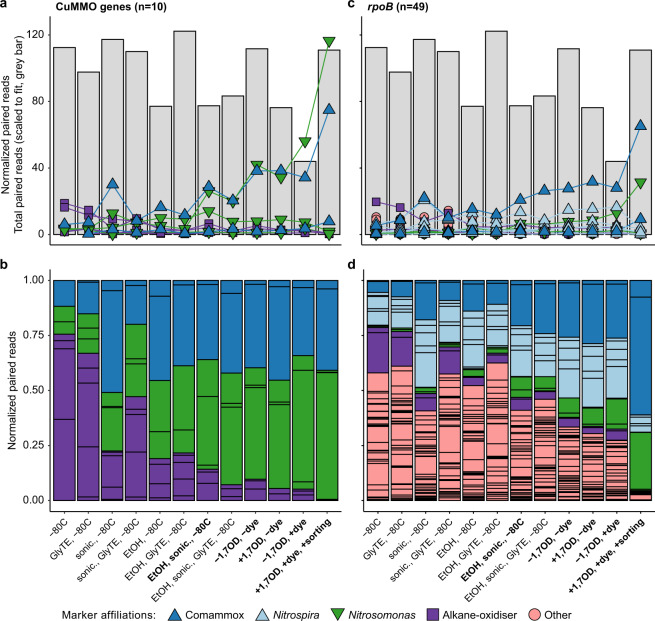
Fig. 7. Targeted metagenomics of CuMMO-containing bacteria.
Normalized read abundances of a, b CuMMO genes and c, d rpoB in the nitrifying enrichment culture of untreated biomass, controls, and sorted biomass for the activity-based labeling protocol (see Supplementary Table S2 for details). Gene abundances were calculated by normalizing paired read counts by gene length and million reads mapped to the co-assembly. The total number of reads mapped to the co-assembly are indicated as gray bars scaled to fit in the same range as the normalized abundances. Colors denote the taxonomic affiliation of the marker genes. The same genes for the taxa of interest (Nitrosomonas, comammox Nitrospira, and putative alkane oxidizers) are emphasized and connected with lines. b, d show the same data as (a, c) but as proportion of the total normalized read abundance for the respective gene in the metagenomic sample, with each individual colored box representing the relative abundance of a single gene from the co-assembly. The final treatment preceding the CuAAC reaction, the CuAAC controls for the influence of 1,7OD inactivation and dye addition, and the final sorted sample are indicated in bold.