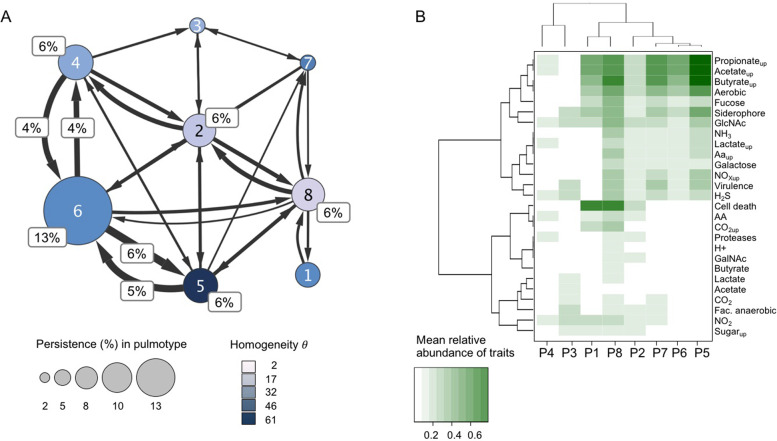
Fig. 3. Dynamic and functional characterization of pulmotypes.
A Transition frequencies between pulmotypes (1–8), proportional (%) representation. Transitions were determined using a sliding window approach within patient trajectories. Patient-stratified counting was applied and average values across all calculations were displayed. Node sizes were scaled according to persistence in the same; edge sizes varied according to frequency of transitions between pulmotypes. Edges supported by <3 transitions were removed; transitions that amount to >3% of total were additionally indicated. Node color represents the DMM cluster homogeneity θ of samples associated with the same pulmotype. B Clustering of pulmotypes by their cross-feeding profiles. Cross-feeding potential was approximated by the mean relative abundance of organisms capable of uptake or production for the respective metabolite. Pulmotype trait vectors were hierarchically clustered after transforming Spearman rank correlation to distances. Trait production/uptake are indicated as name/nameup, respectively. AA amino acids, Cell death reports on apoptosis, necrosis, GlcNAc N-acetylglucosamine, GalNAc N-acetylgalactosamine, NOx nitrate, nitrite, Proteases proteases and elastases, Virulence LPS, ExoS, ExoA, cell toxins, endo toxins, hemolysin.