Abstract
Efficient nutrient cycling in the coral-algal symbiosis requires constant but limited nitrogen availability. Coral-associated diazotrophs, i.e., prokaryotes capable of fixing dinitrogen, may thus support productivity in a stable coral-algal symbiosis but could contribute to its breakdown when overstimulated. However, the effects of environmental conditions on diazotroph communities and their interaction with other members of the coral holobiont remain poorly understood. Here we assessed the effects of heat stress on diazotroph diversity and their contribution to holobiont nutrient cycling in the reef-building coral Stylophora pistillata from the central Red Sea. In a stable symbiotic state, we found that nitrogen fixation by coral-associated diazotrophs constitutes a source of nitrogen to the algal symbionts. Heat stress caused an increase in nitrogen fixation concomitant with a change in diazotroph communities. Yet, this additional fixed nitrogen was not assimilated by the coral tissue or the algal symbionts. We conclude that although diazotrophs may support coral holobiont functioning under low nitrogen availability, altered nutrient cycling during heat stress abates the dependence of the coral host and its algal symbionts on diazotroph-derived nitrogen. Consequently, the role of nitrogen fixation in the coral holobiont is strongly dependent on its nutritional status and varies dynamically with environmental conditions.
Subject terms: Microbial ecology, Climate-change ecology
Introduction
The association with microbial symbionts is central to the ecological success of reef-building corals in the oligotrophic tropical ocean [1, 2]. The close metabolic coupling between heterotrophic corals and their phototrophic algal symbionts supports their immense productivity, has given rise to their rapid radiation, and has led to the formation of coral reef ecosystems [3–7]. Consequently, the destabilization of this symbiosis in times of anthropogenic environmental change is posing a direct threat to the functioning of coral holobionts, i.e., the ecological unit of corals and their associated microorganisms, and the reefs they support [8–10]. In recent years, ocean warming has repeatedly caused mass coral bleaching, the breakdown of the coral-algal symbiosis, frequently followed by the death of the coral host and subsequent ecosystem-wide reef degradation [11, 12]. An in-depth understanding of the processes underlying the functioning and destabilization of the coral-algal symbiosis is thus required to predict or mitigate the effects of climate change on coral reefs [13–16].
Efficient recycling of organic and inorganic carbon in the coral-algal symbiosis depends on the nutrient-limited state of the algal symbionts [17–19]. In a stable state of the symbiosis, low bioavailable nitrogen availability limits algal growth and results in the accumulation of photosynthates in algal cells [20, 21]. The release of these excess nutrients, in turn, fuels the energy metabolism of the coral host. Although low levels of nitrogen assimilation are required to support the holobiont’s net productivity and growth [22], increases in the coral host’s catabolic activity during heat stress or other environmental stressors may increase nitrogen availability for the algal symbionts. Excess nitrogen availability may destabilize nutrient recycling in the coral-algal symbiosis resulting in a breakdown of coral holobiont functioning and eventually host starvation [15, 23–26]. The stability and functioning of the coral holobiont under changing environmental conditions thus depend on its ability to maintain a limited nitrogen availability for the algal symbionts [16, 25].
Importantly, nitrogen assimilation in the coral holobiont is not limited to the coral host and its algal symbionts. In addition to heterotrophic feeding and the uptake of nutrients from seawater, most coral holobionts also show detectable rates of nitrogen fixation, i.e., the prokaryotic conversion of atmospheric dinitrogen into bioavailable ammonium [27–29]. Diazotrophs, Bacteria, and Archaea, capable of nitrogen fixation, are common coral associates and show indications of host specificity [30–34]. As the abundance and activity of diazotrophs are highly dynamic and influenced by environmental change [35–39], nitrogen fixation may either stabilize or destabilize coral holobiont functioning depending on prevailing environmental conditions [25]. During periods of low environmental nitrogen availability, nitrogen fixation activity is positively correlated with coral holobiont productivity and could fulfill up to 11% of the algal symbionts’ nitrogen requirement [38]. In this context, the stimulation of nitrogen fixation rates by temperature stress or eutrophication has been proposed to enhance the productivity and resilience of coral holobionts during stress [40–42]. In addition, stimulated nitrogen fixation and increased nitrogen to phosphorus ratios in the holobiont have been reported during the breakdown of the coral-algal symbiosis under elevated sugar concentrations [43]. The importance of diazotrophs, thus, seems to depend on the fate of fixed nitrogen in the coral holobiont and its effects on other holobiont members [44]. However, our understanding of the environmental drivers controlling nitrogen fixation and the assimilation of diazotroph-derived nitrogen in the coral holobiont remains largely speculative at this point.
Recent studies showed that increased metabolic energy demands and the enhanced release of catabolic waste products may severely alter coral holobiont nutrient cycling during heat stress [15, 45]. We hypothesized that under such conditions, stimulated nitrogen fixation activity would enhance nitrogen availability for algal symbionts and thus contribute further to the destabilization of the coral-algal symbiosis. To test this, here we assessed the effects of heat stress on nitrogen fixation in the coral Stylophora pistillata from the central Red Sea. Combining amplicon sequencing, nutrient flux incubations, isotope labeling, and NanoSIMS imaging, we aimed to assess how heat stress affects diazotroph community structure, nitrogen fixation activity, and the assimilation of diazotroph-derived nitrogen in the coral holobiont before the onset of bleaching.
Materials and methods
Coral collection and experimental design
Five colonies of S. pistillata were collected at Abu Shosha reef (22°18ʹ16.3ʺN; 39°02ʹ57.7ʺE) in the central Red Sea close to the Saudi Arabian coast in September 2018. The colonies were collected at a water depth of ~5 m, at least 10 m apart, and were immediately transported to the indoor aquaria facilities of the Coastal and Marine Resources core laboratory (22°18′20.5″N; 39°06′ 14.3″E) at the King Abdullah University of Science and Technology (KAUST). The experiments for this study were conducted in parallel to those detailed in Rädecker et al. [15] using fragments of the same coral colonies and experimental setup. Coral colonies were fragmented into individual branches of ~5 cm length and nubbins were distributed over two 150 L aquaria per colony (5 colonies × 2 aquaria = 10 aquaria total). Each tank was filled with freshly collected seawater from Abu Shosha reef (salinity = 40.1 ± 0.2; NH4+ = 0.48 ± 0.03 μM; NO3− = 0.19 ± 0.05 μM; PO43− = 0.03 ± 0.00 μM) with a daily water renewal rate of 25%, to maintain stable water parameters inside the aquaria over the course of the experiment (means ± SE for day 10 across aquaria and treatments: salinity = 40.1 ± 0.2; NH4+ = 0.48 ± 0.03 μM; NO3− = 0.19 ± 0.05 μM; PO43− = 0.03 ± 0.00 μM). Seawater nutrient concentrations were measured in three technical replicates per sample using a SA3000/5000 nutrient auto-analyzer (Skalar Analytical B.V, Breda, The Netherlands) according to the manufacturer’s instructions. Each aquarium was equipped with a temperature controller (D-D The Aquarium Solution Ltd, Essex, UK), a 600-W heater (Schego, Offenbach, Germany), a current pump (Tunze, Penzberg, Germany), and a Radion light system (Ecotech Marine, Inc., Bethlehem, PA, USA) to maintain aquaria at a stable temperature of 29.1 °C and a light/dark regime mimicking in situ conditions of the collection site and depth (mean daytime irradiation = 380 μmol quanta m−2 s−1, peak daytime irradiation = 750 μmol quanta m−2 s−1). No supplemental feeding beyond naturally occurring seawater plankton was used in the experiment.
Following 7 days of acclimation, five of the aquaria (one per coral colony) were gradually ramped up to a temperature of 32.9 °C (absolute annual maximum at the collection site in 2017) over the course of 3 days, while the remaining five aquaria were maintained at a constant 29.1 °C (annual mean at the collection site in 2017) [46]. After 7 days at the maximum temperature (i.e., on day 10 of the experiment), replicate fragments from all colonies were sampled in both treatments for all molecular and physiological analyses. For molecular analyses, one nubbin per colony and treatment was immediately flash-frozen in liquid nitrogen and stored at −80 °C until further processing. For physiological analyses, one nubbin per colony and treatment was used for each of the incubations, which were performed immediately after the collection. Investigated response parameters included overall bacterial community composition (16S rRNA gene amplicon sequencing), diazotroph community composition (nifH amplicon sequencing), nitrogen fixation activity (acetylene reduction assay), net assimilation of diazotroph-derived nitrogen (15N2 isotope labeling + bulk analysis), and partitioning of diazotroph-derived nitrogen in the coral holobiont (NanoSIMS imaging of 15N2 isotope assimilation). For details on individual response parameters, see below.
DNA extraction, amplification, and sequencing
Individual coral fragments were placed into Ziploc bags, covered in 0.6 mL of AP1 buffer (Qiagen, Hilden, Germany) and air-blasted to remove their tissue from the coral skeleton using air pressure for 1 min. RNAse A solution (6 µL from a 100 mg/mL stock) was immediately added to the retrieved tissue slurry in AP1 and 400 µL of the resulting slurry was directly used for DNA extraction with the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. In parallel, 400 µL of AP1 buffer + RNase A were used for a no sample (null) DNA extraction to control for possible kit/lab contaminants. The yield and quality of all extractions were assessed using a Nanodrop 2000 (ThermoFischer, Waltham, MA, USA) [47].
For the characterization of the overall prokaryotic community composition, the hypervariable regions V5 and V6 of the 16S rRNA gene were amplified using the primers 784F 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGGATTAGATACCCTGGTA-3′ and 1061R 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCRRCACGAGCTGACGAC-3′ (Illumina overhang adaptor sequences are underlined) [47, 48]. PCRs were run in triplicate using a reaction volume of 10 µL with 5 µL of Qiagen multiplex PCR master mix, 10 ng of DNA template, and final primer concentrations of 0.5 µM for each primer, respectively. PCR cycling conditions consisted of an initial activation step at 95 °C for 15 min, followed by 27 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by a final extension step at 72 °C for 10 min. Successful amplification of all samples was confirmed by gel electrophoresis before products of triplicate PCRs for each sample were pooled. PCR products were subjected to an indexing PCR (8 cycles) using the Nextera XT Index Kit v2 (Illumina, San Diego, USA) according to the manufacturer’s instructions. Indexed samples were then cleaned and normalized using the SequalPrep Normalization Plate Kit (Invitrogen, Carlsbad, CA, USA), pooled at equimolar ratios, and verified on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Amplicon libraries were sequenced at the KAUST Bioscience core laboratory on the MiSeq platform (Illumina) using 2 × 301 bp overlapping paired-end reads with 20% spiked-in phiX.
For the characterization of the diazotroph community composition, the primers IGK3 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGCIWTHTAYGGIAARGGIGGIATHGGIAA-3′ and DVV 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGATIGCRAAICCICCRCAIACIACRTC-3′ (Illumina overhang adaptor sequences are underlined) were used to amplify a region of the nifH gene due to their broad taxonomic range and high specificity as discussed in Gaby and Buckley [49]. PCRs were run in triplicate using a reaction volume of 10 µL with 5 µL of Qiagen multiplex PCR master mix, 10 ng of DNA template, and final concentrations of 1.2 µM for each primer, respectively. PCR cycling conditions consisted of an initial activation step at 95 °C for 15 min, followed by 40 cycles of 95 °C for 45 s, 57 °C for 45 s, and 72 °C for 60 s, followed by a final extension step at 72 °C for 10 min. Successful amplification of all samples was confirmed by gel electrophoresis before products of triplicate PCRs for each sample were pooled and purified using Agencourt AMPure beads (Agencourt Bioscience Corporation, Beverly, MA, USA). Purified PCR products were subjected to an indexing PCR (8 cycles) using the Nextera XT Index Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Indexed samples were then cleaned and normalized using the SequalPrep Normalization Plate Kit (Invitrogen), pooled at equimolar ratios and verified on the Agilent 2100 Bioanalyzer (Agilent Technologies). Amplicon libraries were sequenced at the KAUST Bioscience core laboratory on the MiSeq platform (Illumina) using 2 × 301 bp overlapping paired-end reads with 20% spiked-in phiX.
Sequence processing and analysis
For the 16S rRNA gene-based characterization of the prokaryotic community composition, demultiplexed sequence reads (~54,000 per sample on average) were processed in “DADA2” v.1.18.0 [50] running in “R” v.4.0.3 [51]. Following removal of primer sequences in “DADA2,” reads were quality filtered with a maximum expected error (max EE) of 2, poor quality read ends were trimmed (truncQ = 2), and forward and reverse reads were truncated at 240 and 160 bp, respectively. The error rates were estimated based on all samples and used for inference of true sequence variants. Following the merging of paired reads and removal of chimeras, amplicon sequence variants (ASVs) were classified against the SILVA database version 138 [52] and all non-bacterial ASVs, as well as those with more than 1% relative abundance in negative extractions, were removed from the dataset in “phyloseq” v.1.34.0 [53]. This yielded an average of ~35,000 retained reads per sample distributed over 1,797 ASVs. The effect of the temperature treatment on the bacterial community composition was assessed at the ASV level. For this, the dataset was cleaned from sparsely distributed ASVs using “pime” v.0.1.0 with a prevalence threshold of 40% (~23,000 reads per sample after filtering) and communities were compared using analysis of similarities (ANOSIM) with Bray–Curtis dissimilarities calculated from relative abundances of ASVs as implemented in “vegan” 2.5-7 [54, 55]. Differential abundance of bacterial taxa between temperature treatments was assessed using linear discriminant analysis effect size (LEfSe) as implemented in the “microbial” R package v.0.0.19 [56].
For the nifH-based characterization of the diazotroph community composition, primers were removed from demultiplexed sequence reads (~202,000 per sample) with “Cutadapt” v.2.10 [57]. Using “DADA2” v.1.18.0 [50] running in “R” v.4.0.3 [51], reads were quality filtered with a max EE of 2, poor quality read ends were trimmed (truncQ = 2), and forward and reverse reads were truncated at 220 and 160 bp, respectively. The error rates were estimated based on all samples and used for inference of true sequence variants. Following the merging of paired reads and removal of chimeras, ASVs were translated to the protein level with FrameBot v.1.2.0 using the included nifH reference file [58]. Translated sequences were assigned to nifH gene clusters following Meunier et al. [59], using a consensus assignment based on three criteria adapted after Angel et al. [60]: first, sequences were aligned in “MAFFT” v7.475 [61] and analyzed using a Classification and Regression Tree model following Frank et al. [62]. Second, an Evolutionary Placement Algorithm tree [63] was constructed using the corresponding RaxML implementation and sequences, and were placed on the reference tree after alignment against a reference dataset containing all nifH gene clusters, as implemented in the NifMAP pipeline [60]. Third, sequences were queried against the curated nifH gene database provided by the Zehr lab (https://wwwzehr.pmc.ucsc.edu/nifH_Database_Public) for gene clustering as well as taxonomic assignment. Based on this approach, ASVs assigned to nifH paralogs not involved in nitrogen fixation (clusters IV&V) as well as all ASVs with more than 1% relative abundance in negative extraction were excluded from the dataset in “phyloseq” v.1.34.0 [53]. This yielded an average of ~65,000 retained reads per sample (93,000–186,000 reads including nifH paralogs) distributed over 238 ASVs (876 ASVs including nifH paralogs). The effect of the temperature treatment on the diazotroph community composition was assessed at the ASV level. To do this, the dataset was cleaned from sparsely distributed ASVs using “pime” v.0.1.0 with a prevalence threshold of 60% (~42,000 reads per sample after filtering) and communities were compared using ANOSIM with Bray–Curtis dissimilarities calculated from relative abundances of ASVs as implemented in “vegan” 2.5-7 [46, 47]. Differential abundance of diazotroph taxa between temperature treatments was assessed using LEfSe as implemented in the “microbial” R package v.0.0.19 [56].
Acetylene reduction assays
Nitrogen fixation rates of coral holobionts were quantified indirectly using the acetylene (C2H2) reduction assay [64]. For this, corals were transferred into 1 L glass incubation chambers (Weck, Wehr-Öflingen, Germany) and submerged in filtered seawater (0.22 µm) to a final volume of 720 mL. After this, 80 mL of C2H2-saturated seawater (continuously bubbled with C2H2 for 30 min prior to incubations) was added and the chambers were sealed gas-tight with a modified glass lid containing a syringe injection port. Ten percent (20 mL) of the gas headspace was replaced with pure acetylene. Incubation chambers were transferred into temperature-regulated water baths equipped with magnetic stirrers and Radion lights (Ecotech Marine, Inc.) to maintain temperature and light conditions identical to treatment and aquaria conditions. At the beginning and after 24 h of incubation, 2.5 ml gas samples were collected from the chamber headspace with a gas-tight syringe equipped with a push-pull valve (Trajan, Ringwood, Australia) and transferred into vacuum blood collection tubes for subsequent analysis. Ethylene (C2H4) concentrations in the gas samples were determined using gas chromatography with a helium pulsed discharge detector (Agilent 7890B GC system with HP-Plot/Q column; lower detection limit for C2H4: 0.3 p.p.m.). Rates of C2H4 evolution during the incubations were calculated based on changes in C2H4 concentrations in the chamber taking into account the temperature-dependent solubility of C2H4 in seawater [65] as well as background fluxes of seawater controls without corals to account for possible planktonic background C2H4 production (ca. an order of magnitude lower than in coral incubations) [65]. C2H4 evolution rates were converted to their equivalent nitrogen fixation rates using the theoretical conversion factor of 4 and normalized to the surface area of coral fragments [66]. Importantly, the choice of the correct conversion factor is widely debated and may depend on the species and environmental context, as such absolute rates should be interpreted with caution [67]. For rate normalization, surface areas of coral fragments were estimated by 3D computer modeling [68]. Forty to 50 photos were taken of the coral fragments from all angles. The Autodesk Photo-to-3D cloud service (Autodesk, Mill Valley, CA, USA) was used to generate 3D models, which were used for surface area analysis with ReCap Photo v.4.2.0.2 (Autodesk).
Isotope labeling and bulk isotope analysis
To quantify the assimilation of microbially fixed nitrogen in the coral holobiont, an isotope labeling approach with 15N2 isotope-enriched seawater was adapted after the dissolution technique outlined in Wilson et al. [69]. First, filtered seawater (0.22 µm) was degassed by constant stirring at low pressure (<200 mbar) for 30 min and degassed seawater was transferred into 1 L borosilicate bottles with gas-tight rubber septa. To allow for higher 15N2 concentrations during incubations without depleting oxygen (O2) levels, bottles were injected with 25 mL of a gas mixture containing 80% 15N2 (98 atom%, Cambridge Isotope Laboratories, Tewksbury, MA, USA) and 20% O2 using a gas-tight syringe equipped with a push-pull valve (Trajan). Bottles were vigorously shaken until the bubble volume remained stable and the bottles were stored under constant agitation at 4 °C overnight. 15N2-enriched seawater and freshly filtered seawater (0.22 µm) were mixed at equal parts for the final incubations. Notably, this approach did not compensate for the reduction of pCO2 during the degassing. As such, we cannot rule out that alterations in seawater carbonate chemistry (albeit consistent across all treatments) may have had minor effects on coral physiology during the incubations. For the isotope labeling, corals were incubated in bubble-free, sealed 1 L glass chambers (Weck) with gas-tight glass lids. Incubation chambers were transferred into temperature-regulated water baths equipped with magnetic stirrers and Radion lights (Ecotech Marine, Inc.) to maintain temperature and light conditions identical to treatment and aquaria conditions. After 24 h, the seawater in the incubations was replaced with equal parts of 15N2-enriched seawater and freshly filtered seawater (0.22 µm), and the chambers were sealed again for another 24 h of incubation. After a total of 48 h, incubations were terminated and corals were sampled for isotope analysis. The tip of coral fragments (~0.5 cm) was removed using a clipper and processed for NanoSIMS analyses as outlined below. The remaining fragment was transferred into a ziplock bag, covered in 2 mL of deionized water, and its tissue was removed from the coral skeleton using air pressure. The resulting tissue slurry was freeze-dried and homogenized subsamples were transferred into tin capsules for bulk N isotope analysis. Total nitrogen content, as well as 15N/14N ratios, were measured using an elemental analyzer (Thermo Flash EA 1112) coupled to a stable isotope mass spectrometer (IRMS, DELTA V Advantage). Notably, we did not quantify concentrations of 15N2 in the incubation water. Hence, 15N/14N ratios do not allow for absolute quantification of nitrogen assimilation rates in the present study. However, as all corals were incubated in the same incubation water with identical 15N2 concentrations, levels of 15N enrichments provide a direct reflection of relative nitrogen assimilation rates in these corals. For this, levels of 15N enrichment were expressed using the delta (δ) notation in units per mil of stable isotope ratios calculated as: δ15N (‰) = (15N/14Nlabeled sample/15N/14Nunlabeled sample − 1) × 1,000.
NanoSIMS imaging
Following 15N2 incubations, tips of coral fragments were immediately transferred into a fixative solution (1.25% glutaraldehyde and 0.5% paraformaldehyde in 0.1 M phosphate buffer) at 4 °C for 24 h. Fixed samples were washed once in phosphate-buffered saline (1×), decalcified in 0.1 M ethylenediaminetetraacetic acid (exchanged daily) for 2 weeks after which the tissue was dissected into a strip of two to three individual polyps. The tissue was dehydrated in a series of increasing ethanol concentrations (50%, 70%, 90%, and 100%), transferred to acetone, and gradually infiltrated with increasing concentrations of SPURR resin (25%, 50%, 75%, and 100%). Subsequently, tissues were embedded in SPURR resin and cut into 170 nm sections using an Ultracut E microtome (Leica Microsystems, Wetzlar, Germany) and mounted on pulse-discharge silicon wafers. Wafers with sample sections were gold-coated (ca. 15 nm) and analyzed with the NanoSIMS 50 ion probe [70] at the Center for Microscopy, Characterization, and Analysis at the University of Western Australia. Surfaces of samples were bombarded with a 16 keV primary Cs+ beam focused to a spot size of about 100 nm with a current of ~2 pA. Secondary molecular ions 12C12C−, 12C13C−, 12C14N−, and 12C15N− were simultaneously collected in electron multipliers at a mass resolution (M/ΔM) of about 8,000. Images for all targeted secondary molecular ions were collected by rastering the primary beam across the sample with a dwell time of 9 ms per pixel; six planes were recorded for each area. Charge compensation was not necessary. Although samples from all coral colonies were analyzed on the NanoSIMS, only data and images from the colony with the highest 15N enrichment level in its tissue (including both control and heat stress conditions) are shown here. For this colony, at least 25 images across the polyp tissue (30 μm raster with 256 × 256 pixels) were collected from both treatments, respectively. To avoid any biases between samples due to differences in analyzed tissue areas within the polyp, only images containing both host tissue and algal symbiont cells were included in the analysis. Following this criterion, 15 images from the control as well as the heat treatment were used for the comparison of 15N/14N ratios, respectively. Images were processed with the ImageJ plugin OpenMIMS (National Resource for Imaging Mass Spectrometry, https://github.com/BWHCNI/OpenMIMS/wiki). After drift correction, the individual planes were summed and the 12C14N− maps were used to draw two regions of interest (ROI) per image containing either all host gastrodermis or all algal symbionts cells and the δ15N of each ROI was calculated as outlined above.
Statistical analyses of nitrogen fixation activity and assimilation rates
All statistical analyses were performed in “R” v.4.0.3 [51]. Differences in nitrogen fixation rates (C2H2 reduction), as well as 15N2 assimilation (IRMS), were analyzed in a paired design based on colony replicates using a sign test. The relationship between nitrogen fixation rates and 15N2 assimilation was assessed via a correlation analysis using Pearson’s product–moment correlation coefficient. NanoSIMS measurements of δ15N in the host and symbiont tissue/cells were analyzed using a two-way analysis of variance using holobiont compartment and treatment as explanatory variables.
Results and discussion
Diazotroph community composition varies with temperature
To assess the role of diazotrophs in the early response of the coral holobiont to heat stress, we sampled the colonies of S. pistillata on day 10 of the experiment (7 days at a maximum temperature of 32.9 °C). At this time point, corals from heat stress and control treatments maintained a healthy appearance, similar levels of algal symbiont densities, and showed no visual signs of bleaching (for an extended discussion of processes and timeframes of bleaching during the experiment, please refer to Rädecker et al. [15]). The early heat-stress response did not affect overall bacterial community composition (ANOSIM, p = 0.145; Fig. S1). Members of the order Oceanospirillales, which are prevalent in healthy corals and S. pistillata in particular [48, 71, 72], represented the largest component of the coral microbiomes from both treatments (~20% of 16S rRNA sequences).
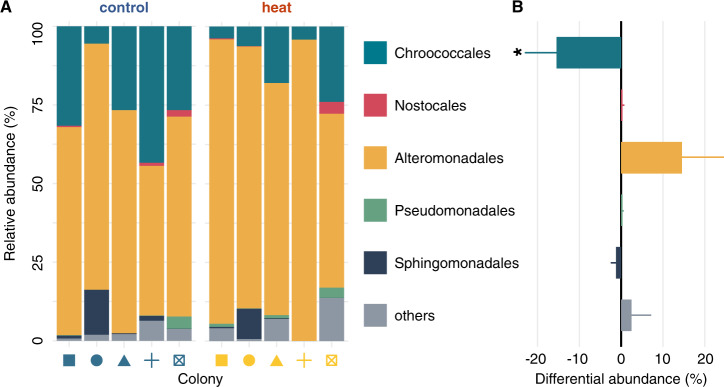
In contrast to the stable overall bacterial microbiome composition, nifH sequencing revealed that heat stress caused a distinct shift in the community composition of diazotrophs (ANOSIM, p = 0.049). This shift was not driven by the loss or the recruitment of novel taxa but rather by variations in the relative abundance of taxa already present in the holobiont (Fig. 1A, B). The diazotroph community was largely dominated by the orders Alteromonadales and Chroococcales accounting for 65% and 27% of nifH sequences in colonies from the ambient control, respectively. Heat stress, however, caused a significant decline in the relative abundance of Chroococcales (LEfSe, χ2 = 3.94, p = 0.047) accompanied by the proportional (albeit not significant) increase of Alteromonadales in the diazotroph community (LEfSe, χ2 = 2.46, p = 0.117) (Fig. 1B). Importantly, Chroococcales were previously identified as endosymbionts in the epithelium of the Caribbean coral Montastrea cavernosa [73] and have been proposed to be an important source of fixed nitrogen for algal symbionts in these corals [74]. Consequently, the observed shifts in diazotroph community composition could directly affect nitrogen availability for other holobiont members during heat stress.
Fig. 1. Characterization of coral-associated diazotroph communities after 10 days of heat stress.
A Relative diazotroph community composition of individual colonies under control (left) and heat stress (right) conditions based on nifH amplicon sequencing. Notably, no archaeal diazotrophs could be detected in the present study. B Change in the relative abundance of dominant diazotroph orders during heat stress relative to control conditions (mean ± SE). Asterisks indicate a significant change (p < 0.05) under heat stress compared to control corals.
Algal symbionts assimilate diazotroph-derived nitrogen in a stable coral holobiont
Using the acetylene reduction technique, we estimated daily nitrogen fixation rates in S. pistillata holobionts. Under ambient control conditions (i.e., 29.1 °C), coral holobionts showed detectable rates of nitrogen fixation around 1.0 ± 0.8 nmol N2 cm−2 day−1 in line with rates previously reported for corals from this region (Fig. 2A) [31, 75, 76]. Further, 15N2 isotope labeling resulted in enriched δ15N values in the soft tissue of coral holobionts (mean ± SE = 16.0 ± 4.7‰), which positively correlated with nitrogen fixation rates across coral colonies (Pearson’s correlation, r = 0.99, p = 0.002, Fig. 2B, C). Although these nitrogen fixation rates may be relatively low compared to other nutrient sources, they are a non-negligible component of the overall nutrient cycling in the nitrogen-limited S. pistillata holobiont.
Fig. 2. Nitrogen fixation activity and assimilation in the coral holobiont after 10 days of heat stress.

A Nitrogen fixation activity of coral holobionts (n = 5 per treatment) was quantified via the acetylene reduction assay. B Net assimilation of diazotroph-derived nitrogen in the coral holobiont (n = 5 per treatment) quantified by 15N2 isotope labeling and bulk stable isotope analysis. C Nitrogen fixation activity and assimilation of diazotroph-derived nitrogen show a positive correlation across colonies (symbols) under control (blue) and heat stress conditions (red) with corresponding confidence intervals (gray). Pearson’s correlation coefficients (r) correspond to correlations among each condition of the corresponding color. Samples from both control and heat stress conditions of the colony with the highest enrichment (ellipse) were imaged in detail using the NanoSIMS ion microprobe. Bars and error bars indicate mean ± SE. Asterisks indicate significant differences between treatments (p < 0.05).
To identify which holobiont partner(s) assimilated diazotroph-derived nitrogen in the coral holobiont, we used NanoSIMS imaging to trace the incorporation of the 15N2 isotope marker in the coral colony with the highest nitrogen fixation activity (Fig. 3A–D). Overall, the algal symbionts showed significantly higher levels of 15N enrichment than the coral host tissue (118.7 ± 9.4‰ (symbionts), 9.5 ± 1.6‰ (host); Tukey’s honestly significant difference (HSD), p < 0.001, Fig. 3F). These observations are consistent with previous reports for S. pistillata from the Great Barrier Reef and show that a large fraction of diazotroph-derived nitrogen is eventually assimilated by the algal symbionts [33]. In addition, NanoSIMS images revealed the presence of small 15N enrichment hotspots in the host epithelium that displayed two distinct morphologies: oval compartments/cells 2–3 µm in length and clusters of smaller circular and rod-shaped compartments/cells with 1–3 µm in length (Fig. 3B, C). Although the NanoSIMS images do not allow us to elucidate the identity of these hotspots, their shape and location are broadly consistent with previous reports of the presence of endosymbiotic bacteria in the coral holobiont [73, 77–79]. Specifically, the larger oval hotspots resemble observations of Chroococcales in the epithelium of M. cavernosa, whereas the rod-shaped clusters resemble previously described bacterial aggregates in the coral tissue in their structure and localization [73, 77–79]. It is thus plausible that the observed epithelial 15N hotspots represent endosymbiotic diazotrophs in the tissue of S. pistillata. However, if these 15N hotspots are indeed diazotrophs, they are not situated near the algal symbionts that are the primary sink of diazotroph-derived nitrogen. In many other phototroph–diazotroph symbioses, diazotrophs predominantly release fixed nitrogen via the passive diffusion of ammonium [80]. Although the ways of nitrogen release by coral-associated diazotrophs are currently unknown, it is plausible that passive ammonium release and/or the catabolic breakdown of diazotrophs during host digestion may contribute to the inorganic nitrogen pool in the holobiont. If this is the case in S. pistillata, the uneven partitioning of diazotroph-derived nitrogen between the coral host and its algal symbionts likely reflects the respective metabolic demands of symbiotic partners for external nitrogen sources. In this scenario, the nitrogen-limited state of algal symbionts might support strong concentration gradients in the host tissue, which would enable efficient assimilation of exogenous nitrogen (derived from either diazotrophs or from the surrounding seawater) [81]. In a stable holobiont state, diazotrophs thus provide a nitrogen source to support algal symbiont growth and allow for net productivity of the coral holobiont (Fig. 4A).
Fig. 3. NanoSIMS imaging of 15N2 assimilation in the coral holobiont after 10 days of heat stress.
A Mosaic of hue saturation images of 15N12C−/14N12C− ratios illustrating the spatial distribution of 15N enrichment in the coral tissue under control conditions. B, C Enlarged 14N12C− NanoSIMS images of epithelial 15N2 hotspots highlighted in A. Epithelial hotspots were primarily characterized by two distinct morphologies: oval compartments/cells with 2–3 µm length (yellow circles) and clusters of smaller rod-shaped compartments/cells with 1–3 µm in length (white circles). D, E Hue saturation images of 15N12C−/14N12C− ratios illustrating 15N assimilation in the coral host and algal symbionts under control (D) and heat stress (E) conditions. F 15N2 assimilation in the coral tissue and algal symbionts under control (blue) and heat stress (red) conditions based on NanoSIMS images (n = 15 per treatment). Differing letters above the boxplot indicate significant differences between groups (p < 0.05). All scale bars are 10 µm.
Fig. 4. Conceptual view of the effect of heat stress on nitrogen assimilation in the coral holobiont.

A In a stable state, low environmental nitrogen availability limits nitrogen uptake in the coral holobiont. Under these conditions, diazotroph-derived nitrogen provides an important nitrogen source for the coral-algal symbiosis. B During heat stress, the release of excess nitrogen waste from the host metabolism increases nitrogen availability in the coral holobiont. Under these conditions, the coral-algal symbiosis does not depend on diazotroph-derived nitrogen. The color of arrows indicates the proportion of diazotroph-derived nitrogen in the nitrogen flux/pool: continuous scale ranging from “low contribution of diazotroph-derived nitrogen (dark blue)” on one end of the spectrum to “high contribution of diazotroph-derived nitrogen (pink)” on the other end of the spectrum. The line width of arrows in the heat stress (B) indicates their proportional increase relative to control conditions (A).
Reduced assimilation of fixed nitrogen by algal symbionts despite increased nitrogen fixation activity during heat stress
On day 10 of the experiment, heat stress caused a 74% increase in nitrogen fixation rates (indirectly quantified as acetylene reduction) compared to coral holobiont rates from ambient control conditions (sign test, z = 2.24, p = 0.025, Fig. 2A). Similar increases in coral-associated nitrogen fixation during heat stress have been previously reported for other coral species [40, 41], suggesting that the here reported processes are unlikely restricted to the S. pistillata holobiont, but may be a common feature of coral-associated diazotroph communities. 15N2 isotope labeling in this study revealed that the additional fixed nitrogen was not assimilated in the soft tissue of the holobiont, in which 15N enrichment decreased by 30% compared to ambient controls (sign test, z = 2.24, p = 0.025, Fig. 2B). NanoSIMS analysis corroborated this observation, revealing that the 15N enrichment remained stable in the host tissue (Tukey’s HSD, p = 0.976) but declined by 53% in the algal symbionts compared to ambient controls (Tukey’s HSD, p < 0.001, Fig. 3E, F). Furthermore, we did not detect any epithelial 15N enrichment hotspots in corals exposed to heat stress, suggesting lower abundances of diazotrophs and/or lower nitrogen fixation activity. The overall increase in nitrogen fixation activity in the coral holobiont during heat stress (Fig. 2A), hence, was likely not driven by endosymbiotic diazotrophs in the coral tissue and did not result in increased assimilation of diazotroph-derived nitrogen by the algal symbionts in the present study (Fig. 3F).
Importantly, the dependence of algal symbionts on diazotroph-derived nitrogen is a function of their nutritional status. In this context, Rädecker et al. [15] recently showed (using the same coral colonies and experimental design) that heat stress shifted these coral holobionts from a state of nitrogen limitation towards a state of carbon limitation. Specifically, energy starvation caused by heat stress promoted the catabolic generation of inorganic nutrients (including ammonium) in the coral tissue, thereby promoting the proliferation of algal symbionts (Fig. S2). This ammonium released by the host catabolism was not isotopically enriched and therefore diluted diazotroph-derived nitrogen in the inorganic nutrient pool. Overall, the reduced 15N enrichment observed here suggests that heat stress reduces the relative contribution of diazotroph-derived nitrogen to the coral holobiont nitrogen pool (Fig. 4B). In this scenario, altered nutrient cycling may help explain the absence of epithelial 15N enrichment hotspots during heat stress. Low photosynthate and high ammonium availability in the coral tissue likely suppress the nitrogen fixation activity of endosymbiotic diazotrophs and may give a competitive advantage to other microbes better adapted to exploit the altered nutrient regime during heat stress [82–84].
It is important to consider that the bulk isotope and NanoSIMS analyses only quantify the assimilation of anabolically incorporated 15N into the coral and symbiont cells. As such, the assimilation of diazotroph-derived nitrogen in other holobiont compartments (i.e., the coral skeleton and the surface mucus layer) is not accounted for in our analyses. Indeed, a study by Moynihan et al. [34] recently suggested that endolithic microbes in the coral skeleton were the main source and sink of diazotroph-derived nitrogen in the closely related Pocillopora acuta holobiont. Further, El-Khaled et al. [85] showed that moderate increases in inorganic nutrient concentrations may stimulate nitrogen fixation rates in Red Sea corals. In this context, the observed net release of ammonium and phosphate by the coral host during heat stress would directly affect nutrient availability in other compartments of the coral holobiont such as the skeleton (Fig. S2 [15]). It is thus plausible that the increase in nitrogen fixation activity described here was predominantly driven by an increase in the activity of endolithic microbes associated with the holobiont. While some studies suggested that endolithic microbial communities may eventually release some of their nutrients to the coral tissue [34, 42, 86, 87], our results clearly show that diazotroph-derived nitrogen (regardless of its origin within the holobiont) is an insignificant source of nitrogen for the coral-algal symbiosis during heat stress. As such, the increase in stimulated nitrogen fixation may primarily be absorbed within the endolithic community itself, thereby supporting the frequently documented rapid proliferation of endolithic microbes during heat stress [86, 88, 89]. Further, nitrogen fixation rates correlate with denitrification activities in some Red Sea corals [75]. Although we did not quantify denitrification rates in the present study, increases in the abundance and or activity of denitrifying microbes may have contributed to the removal of diazotroph-derived nitrogen from the coral holobiont during heat stress. Taken together, we conclude that the excess availability and release of ammonium through host catabolic processes, combined with increased microbial utilization of fixed nitrogen, likely reduce the relative contribution of diazotroph-derived nitrogen to holobiont nitrogen cycling during heat stress.
The functional importance of diazotrophs in the coral holobiont
Nitrogen fixation rates in coral holobionts are highly variable—they can differ between host species, locations, and fluctuate depending on local environmental conditions [34, 37, 38, 41, 42, 82, 87, 90]. Indeed, the molecular, physiological, and ultrastructural characterization of nitrogen fixation presented here paints a complex picture of the role of diazotrophs in coral holobiont functioning. Our results suggest that endosymbiotic diazotrophs in the coral epithelium actively fix nitrogen in the coral holobiont. The characterization of tissue-associated diazotroph communities suggests that the observed 15N hotspots may resemble individual endosymbiotic diazotrophs of the order Chroococcales and bacterial aggregates potentially including diazotrophs of the order Alteromonadales. Further, the disappearance of these 15N hotspots during heat stress coincided with a decline in Chroococcales nifH relative sequence abundance and the reduced 15N assimilation in algal symbiont cells. Taken together, our findings indicate that endosymbiotic diazotrophs in the coral tissue, especially Chroococcales, represent an important source of nitrogen for algal symbionts under nitrogen-limited conditions. Hence, the association with diazotrophs may supplement holobiont nutrition and enable net productivity under oligotrophic conditions [38]. However, this beneficial role of diazotrophs is likely limited to a narrow window of environmental conditions in which algal symbionts are strongly nitrogen-limited. Environmental conditions that reduce the demand for nitrogen in the coral holobiont (e.g., heat stress in the present study), may hence undermine the beneficial role of diazotrophs in holobiont functioning. This dynamic dependence on diazotroph-derived nitrogen in the coral holobiont is directly reflected in the variable diazotroph community assemblage, their activity, and the fate of diazotroph-derived nitrogen in the coral holobiont and may indirectly facilitate holobiont acclimatization and adaptation to changing levels of nutrient availability. In other words, this study suggests that the nutritional status of the coral holobiont determines the structure and activity of associated diazotroph communities and not vice versa. As such, diazotrophs may support coral holobiont functioning under nitrogen-limited conditions. However, anthropogenic impacts such as eutrophication and ocean warming likely reduce the benefits of hosting diazotrophs in the coral holobiont.
Supplementary information
Acknowledgements
We thank Dr Zenon B. Batang and Dr Nabeel M. Alikunhi for their continuous support and assistance with aquaria maintenance. Further, Ioannis Georgakakis, Mustafa Altunkaya, and Professor Matt Kilburn are acknowledged for their help and support with sample processing and data analysis. We thank the editor and the three anonymous reviewers for their constructive feedback and suggestions. NR, CP, AC, MP, J-BR, and CRV were supported by the KAUST competitive research grant URF/1/3400-01-01. CRV also acknowledges funding from the German Research Foundation (DFG), grant 433042944. NR and AM are supported by the Swiss National Science Foundation, grant 200021_179092.
Author contributions
NR, CP, MP, J-BR, and CRV conceived the experiment. NR, CP, HG, and FR conducted the experiment. NR, CP, AC, GP, LG, PG, and JB performed sample and data analysis. All authors contributed to writing and revising the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All raw sequencing data have been deposited under BioProject ID PRJNA741490 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA741490). Further, ASV abundance tables (16S rRNA and nifH gene amplicons) as well as physiological and NanoSIMS data have been deposited at Zenodo.org and are freely available at 10.5281/zenodo.5552980.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01158-8.
References
- 1.Thompson JR, Rivera HE, Closek CJ, Medina M. Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front Cell Infect Microbiol. 2014;4:176. doi: 10.3389/fcimb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pogoreutz C, Voolstra CR, Rädecker N, Weis V, Cardenas A, Raina J-B. The coral holobiont highlights the dependence of cnidarian animal hosts on their associated microbes. In: Bosch TCG, Hadfield MG, editors. Cellular dialogues in the holobiont. CRC Press; 2020. p. 91–118.
- 3.Stanley GD, van de Schootbrugge B. The evolution of the coral–algal symbiosis. In: van Oppen MJH, Lough JM, editors. Coral bleaching: patterns, processes, causes and consequences. Berlin, Heidelberg: Springer; 2009. p. 7–19.
- 4.LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol. 2018;28:2570–80.e6. doi: 10.1016/j.cub.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Muscatine L. The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs. 1990;25:75–87. [Google Scholar]
- 6.Stolarski J, Kitahara MV, Miller DJ, Cairns SD, Mazur M, Meibom A. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol Biol. 2011;11:316. doi: 10.1186/1471-2148-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankowiak K, Wang XT, Sigman DM, Gothmann AM, Kitahara MV, Mazur M, et al. Photosymbiosis and the expansion of shallow-water corals. Sci Adv. 2016;2:e1601122. doi: 10.1126/sciadv.1601122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wild C, Hoegh-Guldberg O, Naumann MS, Colombo-Pallotta MF, Ateweberhan M, Fitt WK, et al. Climate change impedes scleractinian corals as primary reef ecosystem engineers. Mar Freshw Res. 2011;62:205–15. [Google Scholar]
- 9.Hughes TP, Barnes ML, Bellwood DR, Cinner JE, Cumming GS, Jackson JBC, et al. Coral reefs in the Anthropocene. Nature. 2017;546:82–90. doi: 10.1038/nature22901. [DOI] [PubMed] [Google Scholar]
- 10.Kleypas J, Allemand D, Anthony K, Baker AC, Beck MW, Hale LZ, et al. Designing a blueprint for coral reef survival. Biol Conserv. 2021;257:109107. [Google Scholar]
- 11.Anthony KRN, Hoogenboom MO, Maynard JA, Grottoli AG, Middlebrook R. Energetics approach to predicting mortality risk from environmental stress: a case study of coral bleaching. Funct Ecol. 2009;23:539–50. [Google Scholar]
- 12.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science. 2018;359:80–83. doi: 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 13.Voolstra CR, Suggett DJ, Peixoto RS, Parkinson KM, Quigley CB, Silveira M, et al. Extending the natural adaptive capacity of coral holobionts. Nat Rev Earth Environ. 2021;2(11):747–62. doi: 10.1038/s43017-021-00214-3. [DOI] [Google Scholar]
- 14.Suggett DJ, Smith DJ. Coral bleaching patterns are the outcome of complex biological and environmental networking. Glob Chang Biol. 2020;26:68–79. doi: 10.1111/gcb.14871. [DOI] [PubMed] [Google Scholar]
- 15.Rädecker N, Pogoreutz C, Gegner HM, Cárdenas A, Roth F, Bougoure J, et al. Heat stress destabilizes symbiotic nutrient cycling in corals. Proc Natl Acad Sci USA. 2021;118:e2022653118. doi: 10.1073/pnas.2022653118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris LA, Voolstra CR, Quigley KM, Bourne DG, Bay LK. Nutrient availability and metabolism affect the stability of coral–Symbiodiniaceae symbioses. Trends Microbiol. 2019;27:678–89. doi: 10.1016/j.tim.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Muscatine L, Porter JW. Reef Corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience. 1977;27:454–60. [Google Scholar]
- 18.Falkowski PG, Dubinsky Z, Muscatine L, Porter JW. Light and the bioenergetics of a symbiotic coral. Bioscience. 1984;34:705–9. [Google Scholar]
- 19.Falkowski PG, Dubinsky Z, Muscatine L, McCloskey L. Population control in symbiotic corals. Bioscience. 1993;43:606–11. [Google Scholar]
- 20.Peng S-E, Chen C-S, Song Y-F, Huang H-T, Jiang P-L, Chen W-NU, et al. Assessment of metabolic modulation in free-living versus endosymbiotic Symbiodinium using synchrotron radiation-based infrared microspectroscopy. Biol Lett. 2012;8:434–7. doi: 10.1098/rsbl.2011.0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krueger T, Horwitz N, Bodin J, Giovani M-E, Escrig S, Fine M, et al. Intracellular competition for nitrogen controls dinoflagellate population density in corals. Proc R Soc B. 2020;287:20200049. doi: 10.1098/rspb.2020.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberty S, Béraud E, Grover R, Ferrier-Pagès C. Coral croductivity is co-limited by bicarbonate and ammonium availability. Microorganisms. 2020;8:640. doi: 10.3390/microorganisms8050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker DM, Freeman CJ, Wong JCY, Fogel ML, Knowlton N. Climate change promotes parasitism in a coral symbiosis. ISME J. 2018;12:921–30. doi: 10.1038/s41396-018-0046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunning R, Muller EB, Gates RD, Nisbet RM. A dynamic bioenergetic model for coral- Symbiodinium symbioses and coral bleaching as an alternate stable state. J Theor Biol. 2017;431:49–62. doi: 10.1016/j.jtbi.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Rädecker N, Pogoreutz C, Voolstra CR, Wiedenmann J, Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 2015;23:490–7. doi: 10.1016/j.tim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Wiedenmann J, D’Angelo C, Smith EG, Hunt AN, Legiret F-E, Postle AD, et al. Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat Clim Chang. 2012;3:160–4. [Google Scholar]
- 27.Houlbrèque F, Ferrier-Pagès C. Heterotrophy in tropical scleractinian corals. Biol Rev Camb Philos Soc. 2009;84:1–17. doi: 10.1111/j.1469-185X.2008.00058.x. [DOI] [PubMed] [Google Scholar]
- 28.Fiore CL, Jarett JK, Olson ND, Lesser MP. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 2010;18:455–63. doi: 10.1016/j.tim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Grover R, Maguer J-F, Allemand D, Ferrier-Pagès C. Uptake of dissolved free amino acids by the scleractinian coral Stylophora pistillata. J Exp Biol. 2008;211:860–5. doi: 10.1242/jeb.012807. [DOI] [PubMed] [Google Scholar]
- 30.Lema KA, Willis BL, Bourne DG. Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl Environ Microbiol. 2012;78:3136–44. doi: 10.1128/AEM.07800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. Nitrogen fixation aligns with nifH abundance and expression in two coral trophic functional groups. Front Microbiol. 2017;8:1187. doi: 10.3389/fmicb.2017.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson ND, Lesser MP. Diazotrophic diversity in the Caribbean coral, Montastraea cavernosa. Arch Microbiol. 2013;195:853–9. doi: 10.1007/s00203-013-0937-z. [DOI] [PubMed] [Google Scholar]
- 33.Lesser MP, Morrow KM, Pankey SM, Noonan SHC. Diazotroph diversity and nitrogen fixation in the coral Stylophora pistillata from the Great Barrier Reef. ISME J. 2018;12:813–24. doi: 10.1038/s41396-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moynihan MA, Goodkin NF, Morgan KM, Kho PYY, dos Santos AL, Lauro FM, et al. Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef. ISME J. 2021; 10.1038/s41396-021-01054-1. [DOI] [PMC free article] [PubMed]
- 35.Tilstra A, Pogoreutz C, Rädecker N, Ziegler M, Wild C, Voolstra CR. Relative diazotroph abundance in symbiotic Red Sea corals decreases with water depth. Front Mar Sci. 2019;6:372. [Google Scholar]
- 36.Bednarz VN, Cardini U, van Hoytema N, Al-Rshaidat MMD, Wild C. Seasonal variation in dinitrogen fixation and oxygen fluxes associated with two dominant zooxanthellate soft corals from the northern Red Sea. Mar Ecol Prog Ser. 2015;519:141–52. [Google Scholar]
- 37.Rädecker N, Meyer FW, Bednarz VN, Cardini U, Wild C. Ocean acidification rapidly reduces dinitrogen fixation associated with the hermatypic coral Seriatopora hystrix. Mar Ecol Prog Ser. 2014;511:297–302. [Google Scholar]
- 38.Cardini U, Bednarz VN, Naumann MS, van Hoytema N, Rix L, Foster RA, et al. Functional significance of dinitrogen fixation in sustaining coral productivity under oligotrophic conditions. Proc R Soc B. 2015;282:20152257. doi: 10.1098/rspb.2015.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bednarz VN, van de Water JAJM, Grover R, Maguer J-F, Fine M, Ferrier-Pagès C. Unravelling the importance of diazotrophy in corals – combined assessment of nitrogen assimilation, diazotrophic community and natural stable isotope signatures. Front Microbiol. 2021;12:1638. doi: 10.3389/fmicb.2021.631244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos HF, Carmo FL, Duarte G, Dini-Andreote F, Castro CB, Rosado AS, et al. Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J. 2014;8:2272–9. doi: 10.1038/ismej.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardini U, van Hoytema N, Bednarz VN, Rix L, Foster RA, Al-Rshaidat MMD, et al. Microbial dinitrogen fixation in coral holobionts exposed to thermal stress and bleaching. Environ Microbiol. 2016;18:2620–33. doi: 10.1111/1462-2920.13385. [DOI] [PubMed] [Google Scholar]
- 42.Bednarz VN, van de Water JAJM, Rabouille S, Maguer J-F, Grover R, Ferrier-Pagès C. Diazotrophic community and associated dinitrogen fixation within the temperate coral Oculina patagonica. Environ Microbiol. 2019;21:480–95. doi: 10.1111/1462-2920.14480. [DOI] [PubMed] [Google Scholar]
- 43.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Voolstra CR, Wild C. Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob Chang Biol. 2017;23:3838–48. doi: 10.1111/gcb.13695. [DOI] [PubMed] [Google Scholar]
- 44.Bednarz VN, Grover R, Maguer J-F, Fine M, Ferrier-Pagès C. The assimilation of diazotroph-derived nitrogen by scleractinian corals depends on their metabolic status. mBio. 2017;8:e02058–16. doi: 10.1128/mBio.02058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrou K, Nunn BL, Padula MP, Miller DJ, Nielsen DA. Broad scale proteomic analysis of heat-destabilised symbiosis in the hard coral Acropora millepora. Sci Rep. 2021;11:19061. doi: 10.1038/s41598-021-98548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth F, Rädecker N, Carvalho S, Duarte CM, Saderne V, Anton A, et al. High summer temperatures amplify functional differences between coral‐ and algae‐dominated reef communities. Ecology. 2021;102:e03226. doi: 10.1002/ecy.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, et al. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–62. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaby JC, Buckley DH. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE. 2012;7:e42149. doi: 10.1371/journal.pone.0042149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team. R: a language and environment for statistical computing computer program. Vienna, Austria: R Foundation for Statistical Computing; 2021.
- 52.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. The vegan package. Community Ecol Package. 2007;10:631–7. [Google Scholar]
- 55.Roesch LFW, Dobbler PT, Pylro VS, Kolaczkowski B, Drew JC, Triplett EW. pime: A package for discovery of novel differences among microbial communities. Mol Ecol Resour. 2020;20:415–28. doi: 10.1111/1755-0998.13116. [DOI] [PubMed] [Google Scholar]
- 56.Guo K. microbial: do 16s data analysis and generate figures. R package version 1.14.4. 2021.
- 57.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 58.Wang Q, Quensen JF, 3rd, Fish JA, Lee TK, Sun Y, Tiedje JM, et al. Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio. 2013;4:e00592–13. doi: 10.1128/mBio.00592-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meunier V, Geissler L, Bonnet S, Rädecker N, Perna G, Grosso O, et al. Microbes support enhanced nitrogen requirements of coral holobionts in a high CO2 environment. Mol Ecol. 2021;30:5888–99 . [DOI] [PubMed]
- 60.Angel R, Nepel M, Panhölzl C, Schmidt H, Herbold CW, Eichorst SA, et al. Evaluation of primers targeting the diazotroph functional gene and development of NifMAP – a bioinformatics pipeline for analyzing nifH amplicon data. Front Microbiol. 2018;9:703. doi: 10.3389/fmicb.2018.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank IE, Turk-Kubo KA, Zehr JP. Rapid annotation of nifH gene sequences using classification and regression trees facilitates environmental functional gene analysis. Environ Microbiol Rep. 2016;8:905–16. doi: 10.1111/1758-2229.12455. [DOI] [PubMed] [Google Scholar]
- 63.Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardy RW, Holsten RD, Jackson EK, Burns RC. The acetylene-ethylene assay for N2 fixation: laboratory and field evaluation. Plant Physiol. 1968;43:1185–207. doi: 10.1104/pp.43.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Breitbarth E, Mills MM, Friedrichs G, LaRoche J. The Bunsen gas solubility coefficient of ethylene as a function of temperature and salinity and its importance for nitrogen fixation assays: Bunsen coefficient and N2fixation. Limnol Oceanogr Methods. 2004;2:282–8. [Google Scholar]
- 66.Zehr JP, Montoya JP. Measuring N2 fixation in the field. In: Bothe H, Ferguson SJ, Newton WE, editors. Biology of the nitrogen cycle. Elsevier; 2007. p. 193–205.
- 67.Soper FM, Simon C, Jauss V. Measuring nitrogen fixation by the acetylene reduction assay (ARA): is 3 the magic ratio? Biogeochemistry. 2021;152:345–51. [Google Scholar]
- 68.Lavy A, Eyal G, Neal B, Keren R, Loya Y, Ilan M. A quick, easy and non‐intrusive method for underwater volume and surface area evaluation of benthic organisms by 3D computer modelling. Methods Ecol Evol. 2015;6:521–31. [Google Scholar]
- 69.Wilson ST, Böttjer D, Church MJ, Karl DM. Comparative assessment of nitrogen fixation methodologies, conducted in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2012;78:6516–23. doi: 10.1128/AEM.01146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoppe P, Cohen S, Meibom A. NanoSIMS: Technical aspects and applications in cosmochemistry and biological geochemistry. Geostand Geoanal Res. 2013;37:111–54. [Google Scholar]
- 71.Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, et al. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 2017;11:186–200. doi: 10.1038/ismej.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savary R, Barshis DJ, Voolstra CR, Cárdenas A, Evensen NR, Banc-Prandi G, et al. Fast and pervasive transcriptomic resilience and acclimation of extremely heat tolerant coral holobionts from the northern Red Sea. Proc Natl Acad Sci USA. 2021;118:e2023298118. doi: 10.1073/pnas.2023298118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 74.Lesser MP, Falcón LI, Rodríguez-Román A, Enríquez S, Hoegh-Guldberg O, Iglesias-Prieto R. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar Ecol Prog Ser. 2007;346:143–52. [Google Scholar]
- 75.Tilstra A, El-Khaled YC, Roth F, Rädecker N, Pogoreutz C, Voolstra CR, et al. Denitrification aligns with N2 fixation in Red Sea corals. Sci Rep. 2019;9:19460. doi: 10.1038/s41598-019-55408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rädecker N, Pogoreutz C, Ziegler M, Ashok A, Barreto MM, Chaidez V, et al. Assessing the effects of iron enrichment across holobiont compartments reveals reduced microbial nitrogen fixation in the Red Sea coral Pocillopora verrucosa. Ecol Evol. 2017;7:6614–21. doi: 10.1002/ece3.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ainsworth TD, Fine M, Blackall LL, Hoegh-Guldberg O. Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities. Appl Environ Microbiol. 2006;72:3016–20. doi: 10.1128/AEM.72.4.3016-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van de Water JAJM, Ainsworth TD, Leggat W, Bourne DG, Willis BL, van Oppen MJH. The coral immune response facilitates protection against microbes during tissue regeneration. Mol Ecol. 2015;24:3390–404. doi: 10.1111/mec.13257. [DOI] [PubMed] [Google Scholar]
- 79.Wada N, Ishimochi M, Matsui T, Pollock FJ, Tang S-L, Ainsworth TD, et al. Characterization of coral-associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus. Sci Rep. 2019;9:14662. doi: 10.1038/s41598-019-49651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64:781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 81.Pernice M, Meibom A, Van Den Heuvel A, Kopp C, Domart-Coulon I, Hoegh-Guldberg O, et al. A single-cell view of ammonium assimilation in coral–dinoflagellate symbiosis. ISME J. 2012;6:1314–24. doi: 10.1038/ismej.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shashar N, Cohen Y, Loya Y, Sar N. Nitrogen fixation (acetylene reduction) in stony corals: evidence for coral-bacteria interactions. Mar Ecol Prog Ser. 1994;111:259–64. [Google Scholar]
- 83.Inomura K, Bragg J, Riemann L, Follows MJ. A quantitative model of nitrogen fixation in the presence of ammonium. PLoS ONE. 2018;13:e0208282. doi: 10.1371/journal.pone.0208282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agawin NSR, Rabouille S, Veldhuis MJW, Servatius L, Hol S, van Overzee HMJ, et al. Competition and facilitation between unicellular nitrogen-fixing cyanobacteria and non-nitrogen-fixing phytoplankton species. Limnol Oceanogr. 2007;52:2233–48. [Google Scholar]
- 85.El-Khaled YC, Roth F, Tilstra A, Rädecker N, Karcher DB, Kürten B, et al. In situ eutrophication stimulates dinitrogen fixation, denitrification, and productivity in Red Sea coral reefs. Mar Ecol Prog Ser. 2020;645:55–66. [Google Scholar]
- 86.Fine M, Loya Y. Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc R Soc B. 2002;269:1205–10. doi: 10.1098/rspb.2002.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sangsawang L, Casareto BE, Ohba H, Vu HM, Meekaew A, Suzuki T, et al. 13C and 15N assimilation and organic matter translocation by the endolithic community in the massive coral Porites lutea. R Soc Open Sci. 2017;4:171201. doi: 10.1098/rsos.171201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fine M, Roff G, Ainsworth TD, Hoegh-Guldberg O. Phototrophic microendoliths bloom during coral “white syndrome. Coral Reefs. 2006;25:577–81. [Google Scholar]
- 89.Fine M, Steindler L, Loya Y. Endolithic algae photoacclimate to increased irradiance during coral bleaching. Mar Freshw Res. 2004;55:115–21. [Google Scholar]
- 90.Meunier V, Bonnet S, Benavides M, Ravache A, Grosso O, Lambert C, et al. Diazotroph-derived nitrogen assimilation strategies differ by scleractinian coral species. Front Mar Sci. 2021;8:1018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequencing data have been deposited under BioProject ID PRJNA741490 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA741490). Further, ASV abundance tables (16S rRNA and nifH gene amplicons) as well as physiological and NanoSIMS data have been deposited at Zenodo.org and are freely available at 10.5281/zenodo.5552980.