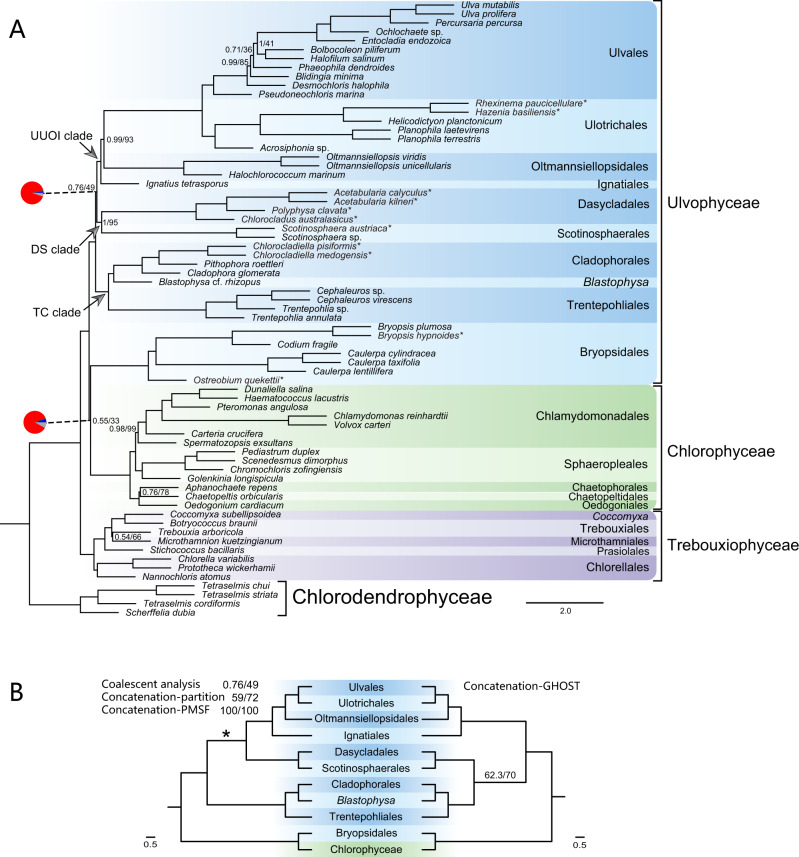
Fig. 2. Phylogenetic relationships of Ulvophyceae inferred from 884 nuclear genes.
A Species tree based on the multispecies coalescent model. Support for each node is provided by local posterior probability and multi-locus bootstrapping (PP/MLBS). Nodes without a value indicate full support. The pie charts at two focal nodes of Ulvophyceae present the proportion of gene tree concordance and conflict. Pie chart color coding: blue, fraction of gene trees that are concordant with the species tree; green, fraction of gene trees supporting the second most common conflicting topology; red, fraction of gene trees supporting all other alternative conflicting partitions; gray, fraction of gene trees with <50% bootstrap support at that node. The newly sampled species are marked with a symbol (*). Note that Desmochloris has been placed in the Chlorocystidales105, and Pseudoneochloris has been recently placed in a new order, Sykidiales106. B Topological comparison among coalescent- and concatenation-based analyses. Support for the conflicting branches is provided by PP/MLBS (in coalescent-based analysis) and SH-aLRT/ultrafast bootstrap (in concatenation-based analyses). The asterisk indicates support in the three analyses on the left. The source data of the pie charts in (A) are provided in the Source Data file.