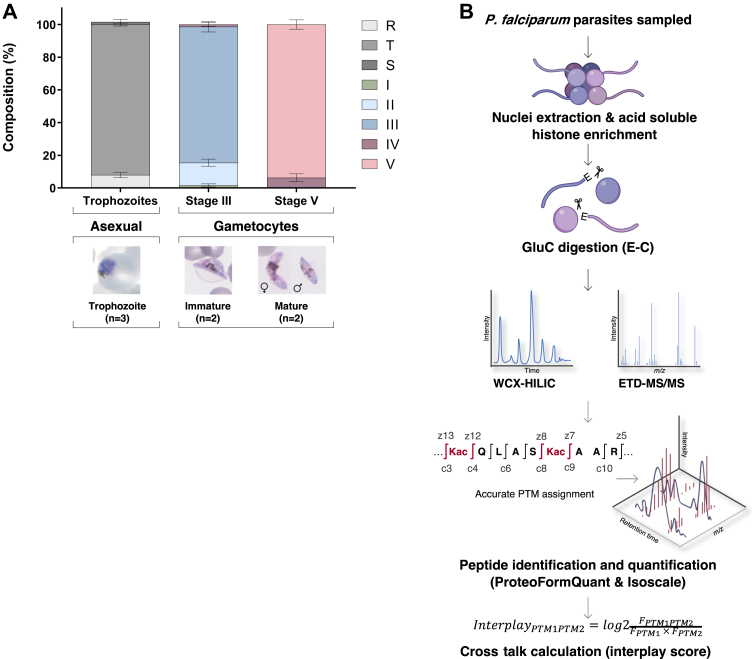
Fig. 1.
Middle-down MS workflow for the analysis of P. falciparum parasite.A, the stage composition of the three biological stages analyzed in this study and representative morphology. Trophozoite (mean ± SEM) samples contained a small percentage of ring (R) and schizont (S) stages, whereas the stage III samples (mean ± SD) contained stage I (I), II (II), III (III), and IV (IV), and stage V samples (mean ± SD) consisted of stage III, IV, and V gametocytes. The trophozoites were from three independent biological repeats and gametocyte stages from two independent biological repeats. B, the middle-down proteomics workflow. Histones were enriched from trophozoite, stage III, and stage V gametocytes and digested using endoprotease GluC, which cleaves at the C terminus of glutamic acid residues, generating intact N-terminal histone H3 peptides (50 amino acid residues in length). To allow sample loading in aqueous buffer and for the most efficient separation for histone N-terminal tails, nano liquid chromatography equipped with a two-column system consisting of a C18-AQ trap column and a weak cation exchange-hydrophilic interaction chromatography (WCX-HILIC) resin analytical column coupled online with high-resolution tandem mass spectrometry (MS-MS) fragmentation was performed using electron transfer dissociation (ETD). Spectra were identified using the Mascot and peptides were quantified using isoScale. The flask image was adapted from the image from Servier Medical Art (http://smart.servier.com/). Servier Medical Art is licensed under a Creative Commons Attribution 3.0 License (CC BY 3.0 license: https://creativecommons.org/licenses/by/3.0/). Data processing and analysis workflow involved MS spectral deconvolution using Xtract (Thermo Fisher Scientific) followed by database searching using Mascot (Matrix Science) with files generated from PlasmoDB (https://plasmodb.org/) and subsequent removal of ambiguously mapped PTMs and stringent quantification (including cofragmented isobaric species) using IsoScale Slim (http://middle-down.github.io/Software). The relative abundance of an individual PTM (PTM1/2) is calculated by summing the relative abundances of all proteoforms carrying the specific individual PTM. The interplay between two individual modifications is calculated by dividing the observed abundance of bivalent PTMs (FPTM1PTM2) with the predicted frequency of a combinatorial PTM (FPTM1 x FPTM2). FPTM1PTM2 is calculated by summing the relative abundances of all proteoforms carrying both PTMs. PTM, posttranslational modification.