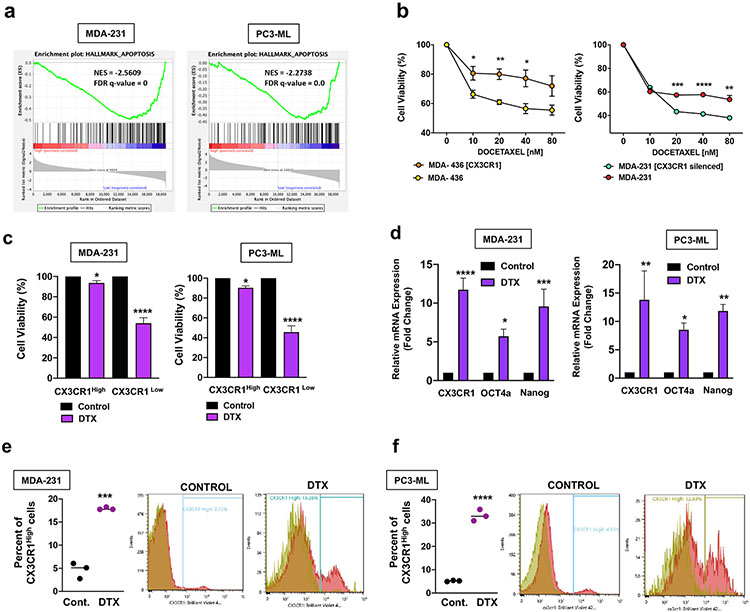
Figure 7.
CX3CR1High cells show resistance to docetaxel. a. GSEA was performed on Hallmark canonical pathways and both the MDA-231 and PC3-ML CX3CR1High sub-populations were de-enriched of the apoptosis signature. b. MDA-436 cells either wild-type or over-expressing CX3CR1 were treated with the indicated doses of docetaxel for 72 hours and assessed for cell viability. MDA-436 cells over-expressing CX3CR1 were significantly less sensitive to docetaxel than wild-type cells. MDA-231 wild-type cells and MDA-231 cells with CX3CR1 silenced by CRISPRi were treated with the indicated doses of docetaxel for 72 hours and cell viability was assessed. MDA-231 cells with CX3CR1 silenced were significantly more sensitive to docetaxel than wild-type cells. c. CX3CR1Low and CX3CR1High sub-populations isolated by FACS from MDA-231 and PC3-ML parental cells were treated with 20 nM docetaxel for 72 hours, and cell viability was compared to the respective untreated population. Both MDA-231 and PC3-ML CX3CR1Low cells showed significantly lower cell viability with docetaxel treatment. d. PC3-ML parental cells were treated with 20 nM docetaxel for 10 days and MDA-231 parental cells were treated with 100 nM docetaxel for 5 days. The surviving populations of cells were collected for quantification of CX3CR1, OCT4a, and NANOG mRNA. Compared to the untreated controls, the MDA-231 and PC3-ML cells treated with docetaxel had significantly higher levels of CX3CR1, OCT4a, and NANOG transcripts. e, f. The MDA-231 and PC3-ML cells that survived docetaxel treatment were collected for analysis of CX3CR1 cell-surface expression by flow cytometry. In comparison to untreated cells, the MDA-231 and PC3-ML cells surviving docetaxel treatment harbored significantly larger CX3CR1High sub-populations. (a: FDR q-value=0.0 and NES=−2.5609 (MDA-231); FDR q-value=0.0 and NES=−2.2738 (PC3-ML); b: *P=0.038; **P=0.003; *P=0.02 (left); ***P=0.0002; ****P<0.0001; **P=0.005 (right); c: *P=0.01; ****P<0.0001; n=6/data point (MDA-231); *P=0.02; ****P<0.0001; n=4/data point (PC3-ML); d: ****P<0.0001; ***P<0.0004; *P=0.0154 (MDA-231); *P=0.04; **P=0.002 (CX3CR1); **P=0.005 (NANOG) (PC3-ML); e: ***P=0.0002; F: ****P<0.0001. One-way ANOVA or Student’s t-test).