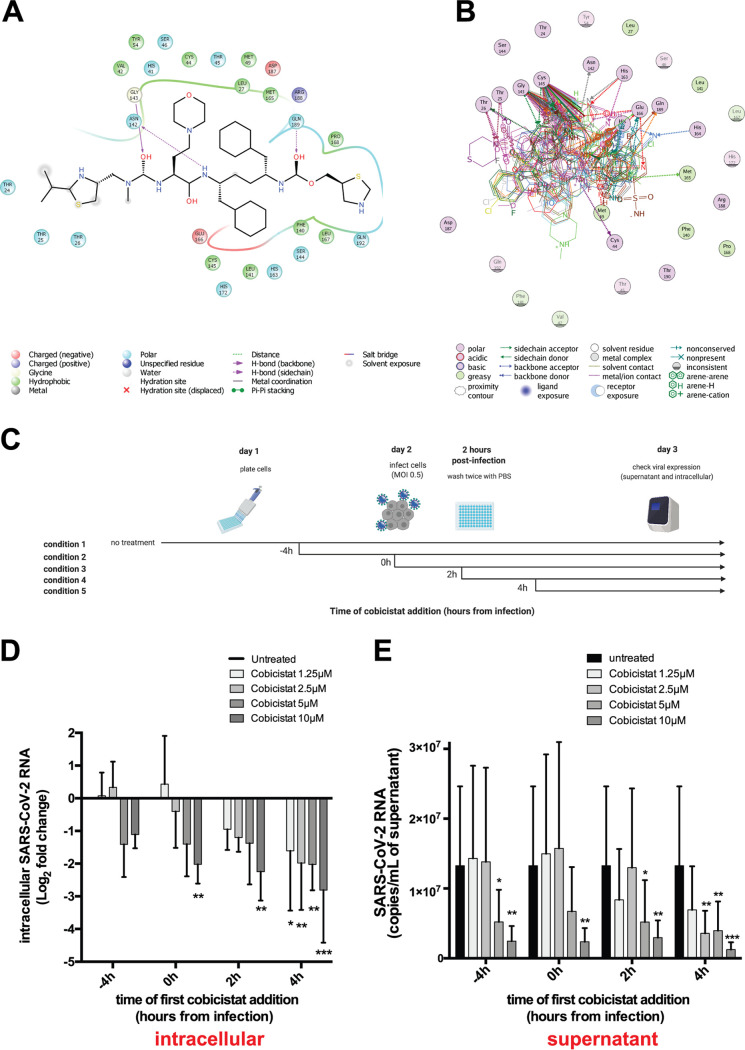
FIG 1.
Cobicistat is a candidate inhibitor of SARS-CoV-2 replication. (A and B) In silico docking analysis of the putative mode and energy of binding of cobicistat to SARS-CoV-2 3CLpro. (A) Docking pose showing the ligand interaction of cobicistat to the active site of 3CLpro and the formation of hydrogen bonds to Asn142, Gly143, and Gln189 of 3CLpro. (B) Overlay of crystal structures of SARS-Cov-2 3CLpro showing the amino acids important for the binding of cobicistat to the active site of the enzyme. Residues of the catalytic dyad (Cys145 and His41) of 3CLpro were among the highest contributors to noncovalent binding to cobicistat. The source and list of structures used are detailed in Materials and Methods. (C) Schematic representation of time course experiments evaluating in vitro inhibition of SARS-CoV-2 replication by cobicistat (created with BioRender). (D and E) Effect of various concentrations of cobicistat, added according to the scheme of panel C, on intracellular and supernatant SARS-CoV-2 RNA content in Calu-3 cells. Viral RNA content was measured by qPCR using the 2019-nCoV_N1 primer set (Centers for Disease Control and Prevention). Fold change values in intracellular RNA (D) were calculated by the delta-delta CT method (74), using the Tata-binding protein (TBP) gene as housekeeper control. Expression levels in supernatant (E) were quantified using an in vitro-transcribed standard curve generated as described in Materials and Methods. Data are expressed as mean with standard deviation (SD) and were analyzed by two-way ANOVA followed by Dunnett’s posttest (n = 3 independent experiments). *, P < 0.05; **, P < 0.01; ***, P < 0.001.