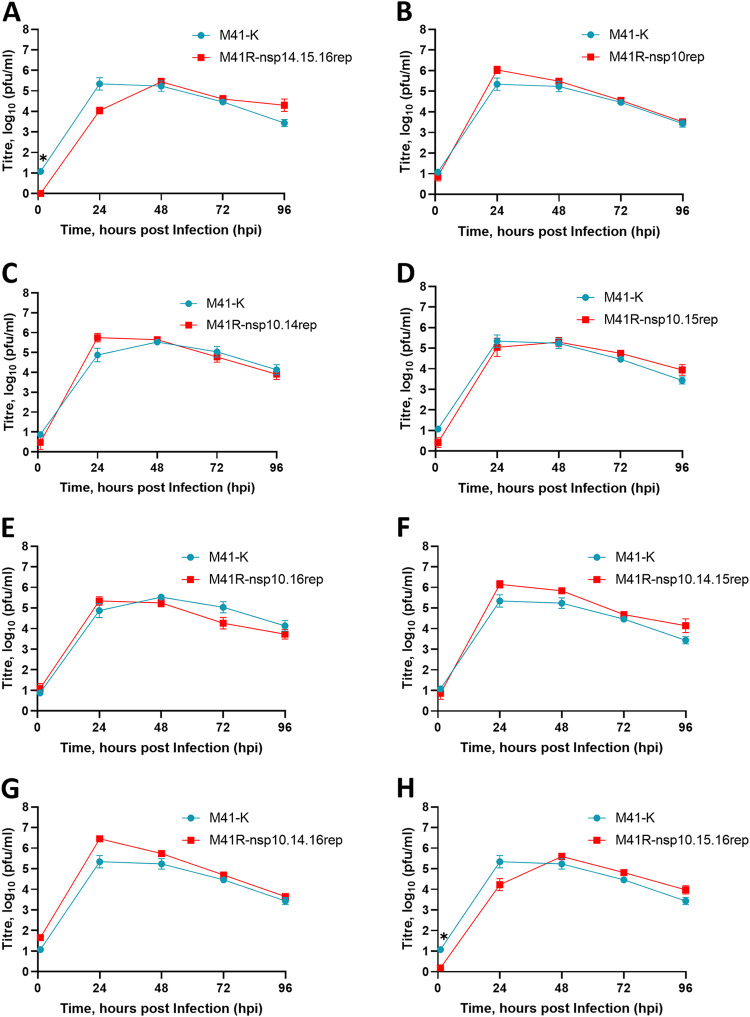
FIG 7.
Combinations of the nucleotides identified at positions 12,137, 18,114, 19,047, and 20,139 in nsps 10, 14, 15, and 16, respectively, do not affect replication in vitro. A series of rIBVs were generated which contained either C/U, G/C, U/A, or G/A at positions 12,137, 18,114, 19,047, and 20,139 within nsps 10, 14, 15, and 16, respectively. All rIBVs were generated through modification of the M41-R backbone to replace (rep) the nucleotides at positions 12,137, 18,114, 19,047, and 20,139 to resemble the pathogenic M41-CK 454 sequence (GenBank accession no. MK728875.1). The IBVs are named according to the nsp location of the nucleotide which was replaced to resemble the latter sequence. Primary CK cells were inoculated with 104 PFU of rIBV M41-K-6 and (A) rIBV M41R-nsp14.15.16rep (B) rIBV M41R-nsp10rep, (C) rIBV M41R-nsp10.14rep, (D) rIBV M41R-nsp10.15rep, (E) rIBV M41R-nsp10.16rep, (F) rIBV M41R-nsp10.14.15rep, (G) rIBV M41R-nsp10.14.16rep, or (H) rIBV M41R-nsp10.15.16rep. Supernatants were harvested at 24-h intervals and the quantities of infectious progeny virus present were determined by plaque assay in CK cells. Each point represents the mean of three independent experiments with error bars representing SEM. Statistical differences were assessed using a two-way ANOVA using Tukey analysis for multiple comparisons and are indicated by * (P < 0.05).