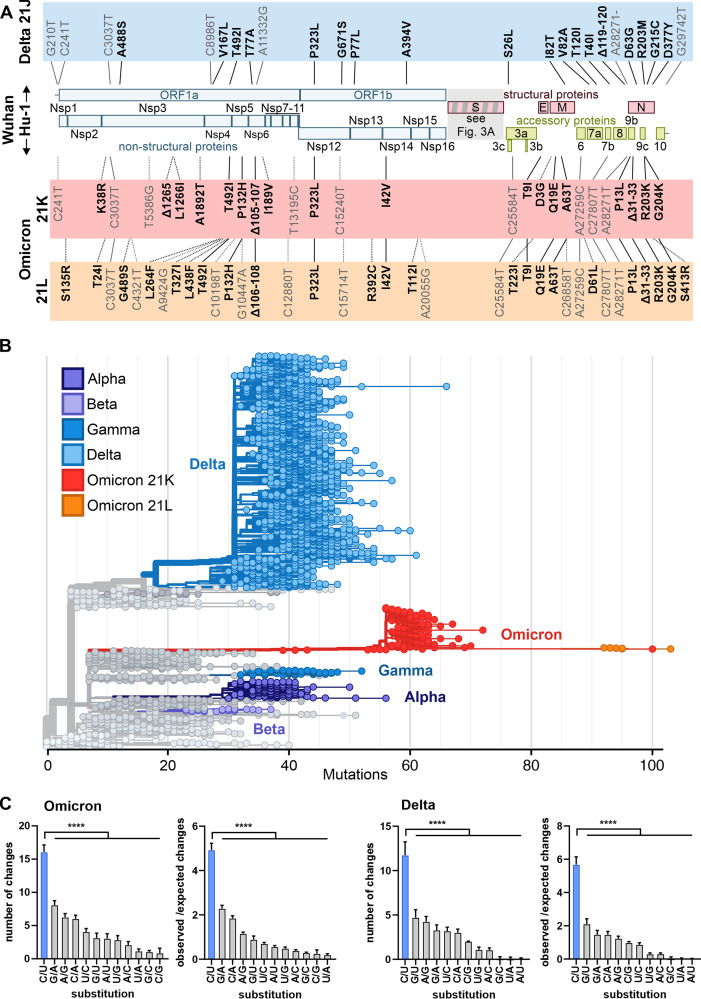
FIG 1.
Features of the Omicron genome and potential consequences of changes outside the Spike gene. (A) Schematic depiction of the SARS-CoV-2 genome and highlighting of non-Spike defining mutations (according to CoVariants data retrieved on 17 January 2022) in the Delta (blue, prevalent 21J clade; https://covariants.org/variants/21J.Delta) and Omicron (red, 21K, also known as BA.1, https://covariants.org/variants/21K.Omicron; orange, 21J, also known as BA.2, https://covariants.org/variants/21L.Omicron; shared mutations are indicated by solid lines) VOCs compared to the Wuhan Hu-1 isolate. Nonsynonymous (bold) and synonymous (nonbold) mutations are indicated. Positions are given relative to the start of the altered protein (nonsynonymous mutations) or the first nucleotide of the genome (synonymous mutations). Nonstructural proteins, Nsp1 to -16 (blue). Structural proteins, Spike (S), Envelope (E), Membrane (M), and Nucleocapsid (N) (red). Accessory proteins, ORF3 to -10 (green). (B) Phylogenetic analysis of representative SARS-CoV-2 isolates scaled according to their divergence compared to the Wuhan Hu-1 sequence. Retrieved from Nextstrain on 18 January 2022 (https://nextstrain.org/ncov/gisaid/global?m=div) and modified. Color coding according to VOC as indicated. (C) Types of single nucleotide substitutions in SARS-CoV-2 Omicron and Delta VOCs compared to Wuhan Hu-1 reference strain. Shown are absolute numbers and ratios of changes observed to changes expected from random mutations based on SARS-CoV-2 nucleotide composition. Sequences were obtained from GISAID and NCBI virus databases (96, 97) and aligned to Wuhan Hu-1 strain (NCBI: NC_045512.2) using Clustal Omega (98). Mean of n = 475 (Delta) or n = 77 (Omicron) + standard deviation. P < 0.0001 (****), one-way paired analysis of variance with multiple comparisons to C/U set.