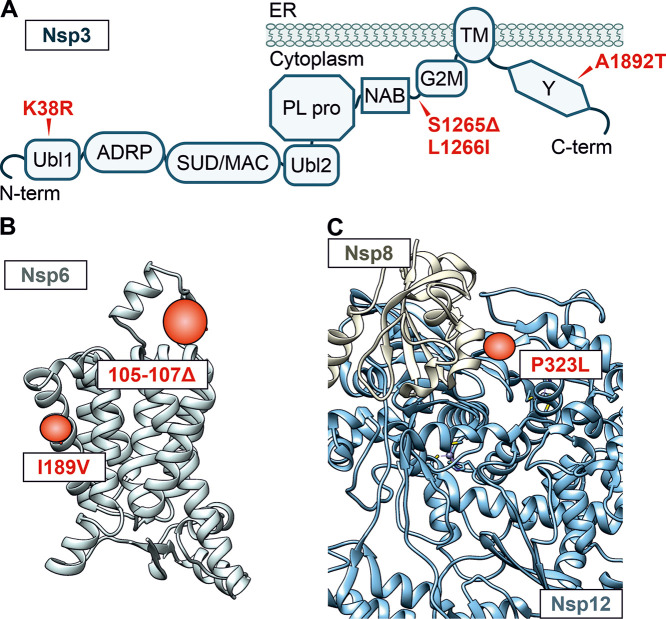
FIG 2.
Alterations in Nsp3, Nsp6, and Nsp8/Nsp12. (A) Schematic depiction of the domain arrangement of Nsp3. Ubl1, ubiquitin-like domain 1; ADRP, ADP-ribose-1″-phosphate phosphatase; SUD/MAC, SARS-CoV unique/macrodomain; Ubl2, ubiquitin-like domain 2; PL pro, papain-like protease; NAB, nucleic acid-binding domain; G2M, group 2-specific marker; TM, transmembrane domain; Y, Y domain. Mutations in the Omicron consensus sequence compared to Hu-1 are highlighted in red, and position is indicated with an arrow. (B) Alpha-fold prediction of the Nsp6 structure. Regions that are altered in the Omicron VOC are highlighted in red. Respective amino acid deletions (positions 105 to 107) or changes (I189V) are labeled in the box. (C) Structural representation of the complex between Hu-1 Nsp8 (tan) and Nsp12 (blue) derived from cryo-EM data (PDB 6YYT). The alteration in the Omicron VOC at the interface is highlighted in red (P323L).