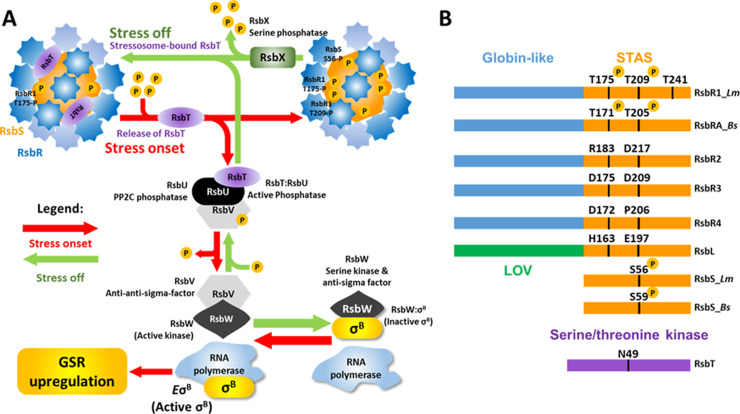
Fig 1. Model depicting the stressosome-mediated regulation of σB and alignment of stressosome proteins.
(A) The putative sensory proteins RsbR1 and its paralogues RsbR2, RsbR3, RsbL, the scaffold protein RsbS and the serine-threonine kinase RsbT form the stressosome. Environmental/nutritional stress is hypothesized to be sensed by one or more RsbR1 paralogue via its N-terminal non-haem binding globin domains forming the turrets at the stressosome surface (blue). The stressosome core (orange) is composed of the STAS domains of both RsbS and RsbR1 C-termini. Stress triggers T209 phosphorylation in RsbR1 and S56 in RsbS, freeing RsbT (purple) from the stressosome core. The phosphatase RsbU interacts with the free RsbT, activating its phosphatase activity towards RsbV. Once dephosphorylated, the serine kinase and anti-sigma factor RsbW, which has higher affinity with the non-phosphorylated RsbV, releases σB and phosphorylates RsbV once more. σB is then able to interact with the RNApol forming the holoenzyme EσB enabling the upregulation of the general stress regulon. The phosphatase RsbX is responsible for the deactivation of the stressosome by dephosphorylating its residues and enabling the re-sequestration of RsbT and the inactivation of the signal cascade. The model shown incorporates the current available data from studies on both Listeria and Bacillus (see text for citations). (B) Domain organization of L. monocytogenes RsbR1_Lm and its paralogues, RsbS_Lm and RsbT and B. subtilis RsbRA_Bs and RsbS_Bs. The N-terminal domains of RsbR1 and paralogues fold into non-haem binding globin-like domains that form the stressosome turrets, while the blue-light sensor RsbL folds into an LOV domain. The C-termini of RsbR1 and paralogues fold into STAS domains forming the stressosome core along with RsbS. The serine-threonine kinase RsbT responsible for the stressosome phosphorylation requires the ATP-Mg2+ chelating residue N49 for its kinase activity.