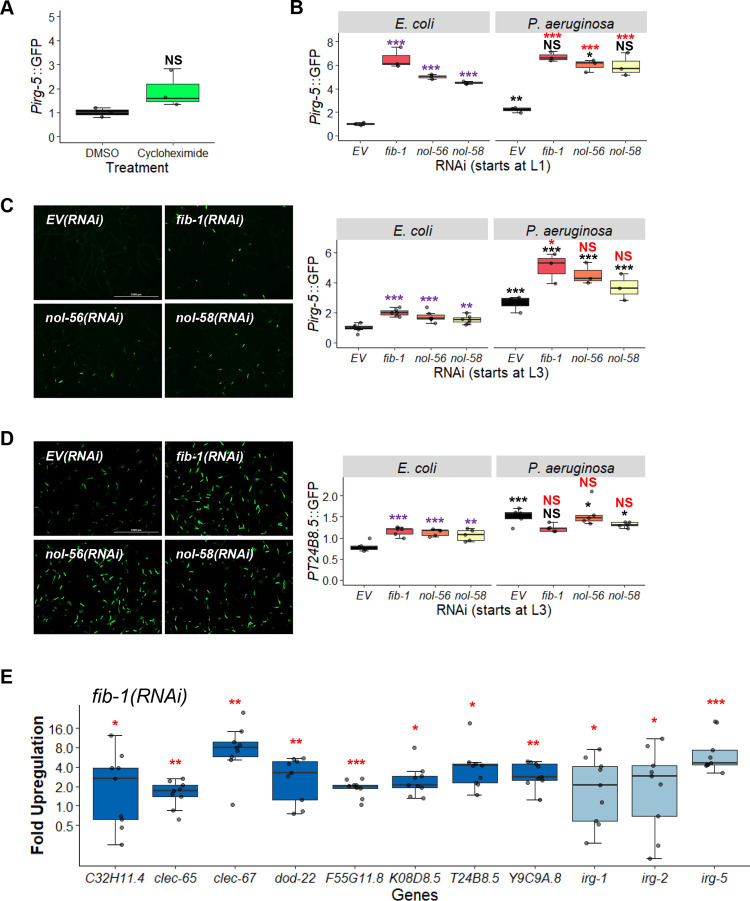
Fig 6. Knockdown of box C/D snoRNPs increases immune activation.
Quantification of GFP fluorescence of C. elegans carrying Pirg-5::GFP (A-C) or PT24B8.5::GFP (D) reporter. (A) Worms were treated for 8 hours with vehicle (DMSO) or translation elongation inhibitor cycloheximide (2 mg/mL). (B-D) Worms were reared on E. coli expressing empty vector (EV), fib-1(RNAi), nol-56(RNAi), or nol-58(RNAi). Young adult worms were transferred onto plates containing E. coli or P. aeruginosa for 8 hours before imaging. RNAi treatment was started at the L1 (B) or L3 (C, D) stage. Representative images are shown on the left for unstressed conditions (C, D). (E) Fold-upregulation of immune genes under the regulation of both UPRmt and p38 MAPK pathways (dark blue) or genes under the control of other regulatory pathways (light blue) in wild-type worms reared on fib-1(RNAi) from the L3 stage. Three biological replicates with ~400 (A-D) or ~8,000 (E) worms/replicate. p-values were determined from two-way ANOVA, followed by Dunnett’s test, or Student’s t-test for GFP quantification assays, or one-way ANOVA, followed by Dunnett’s test for qRT-PCR. All fold-changes were normalized to EV control on E. coli. NS not significant, *p < 0.05, ** p < 0.01, *** p < 0.001. In panels with multi-colored significance marks, purple indicates comparison between gene(RNAi) and EV(RNAi) in the control condition (E. coli), red indicates comparison between gene(RNAi) and EV(RNAi) after exposure to P. aeruginosa, and black indicates comparison between stressed and control conditions.