FIGURE 1:
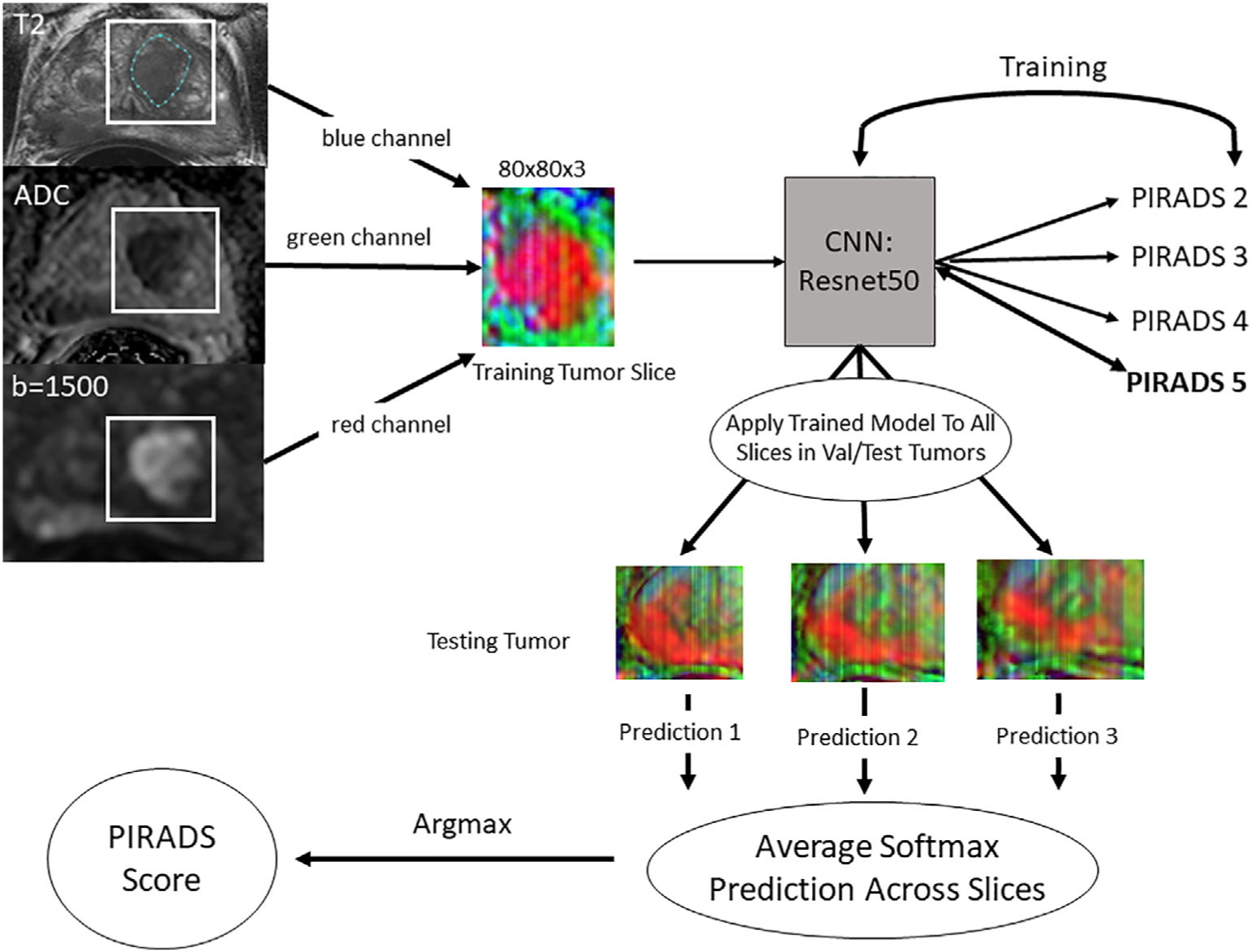
Workflow for data processing, model training, and per-lesion model application. The lesion was segmented on the T2-weighted axial series while viewing the corresponding ADC map and the high-b-value images. All three series were aligned so they were in the same physical space. The lesion segmentation was used to determine the maximum and minimum x and y values, then a bounding box was drawn around the lesion with 10-voxel padding. These cropped images from each series were placed into a three-channel array and saved as JPEGs. All images were resized to 80 × 80. The lesions were then split into 70/20/10 train/validation/test sets and slices were extracted from the training and validation datasets for model training. A Resnet 34 convolutional neural network (CNN) was trained using these slices. The fully trained model was then applied on a slice-by-slice basis for each lesion in the validation and test datasets and the predictions were averaged. The largest average score was then considered the PI-RADS score.
