Abstract
Seven new arylpyrrole alkaloids (1–7), along with four known compounds, were isolated from an extract of a Dactylia sp. nov. marine sponge, and their structures were elucidated by interpretation of NMR and MS spectroscopic data. Denigrins D–G (1–4) have highly substituted pyrrole or pyrrolone rings in their core structures, while dactylpyrroles A–C (5–7) have tricyclic phenanthrene cores. Due to the proton-deficient nature of these scaffolds, key heteronuclear correlations from 1H–15N HMBC and LR-HSQMBC NMR experiments were used in the structure assignment of denigrin D (1). Dictyodendrin F (8), a previously described co-metabolite, inhibited transcription driven by the oncogenic PAX3-FOXO1 fusion gene with an IC50 value of 13 μM.
Graphical Abstract

Rhabdomyosarcoma (RMS) is a soft tissue sarcoma that predominantly occurs in children and young adults. RMS tumors are divided into two subtypes, embryonal and alveolar, based on their histologic features. The PAX3-FOXO1 fusion gene, resulting from reciprocal chromosomal translocations between chromosomes 2 and 13, is a signature genetic change found specifically in the alveolar subtype of rhabdomyosarcoma, and it is not found in other cancers.1 Embryonal RMS and PAX gene fusion-negative alveolar RMS are generally associated with good therapeutic outcomes, whereas PAX gene fusion-positive tumors are more aggressive, often metastatic, and less responsive to chemotherapies and have a worse prognosis. Furthermore, among fusion-positive tumors, patients with the PAX3-FOXO1 translocation generally have worse outcomes compared to those with the PAX7-FOXO1 gene fusion.2 While the need for PAX3-FOXO1 active agents is significant, natural products have not been systematically evaluated as potential therapeutic leads in the past.
In an effort to discover new inhibitors of PAX3-FOXO1 from natural products, an extract of a Dactylia sp. nov. marine sponge was found to inhibit the activity of an engineered PAX3-FOXO1 reporter system during primary high-throughput screening. The genus Dactylia belongs to the Callyspongiidae family, and 14 species have been characterized so far;3 yet few chemical investigations of this genus have been reported.4,5 Herein, we describe the isolation and structure elucidation of seven new alkaloids and four known compounds from a collection of Dactylia sp. nov. that was made in the Maldives. The PAX3-FOXO1 inhibitory activity of all the isolated sponge metabolites was also evaluated.
RESULTS AND DISCUSSION
In a continuation of our prior investigation of this Dactylia sp. sponge sample,5 the organic solvent extract was sequentially chromatographed on a Diol MPLC column and C18 reversed-phase HPLC to yield seven new (1–7) and four known compounds, dictyodendrin F (8), denigrins A and B, and spirodactylone. We previously reported our discovery of the Dactylia metabolite spirodactylone and its generation from denigrin B, which was a co-metabolite in the extract.5
Denigrin D (1) was obtained as a yellow amorphous solid, and the (+)-HRESIMS spectrum displayed a protonated molecule [M + H]+ at m/z 600.2395, corresponding to a molecular formula of C38H33NO6 with 23 degrees of unsaturation. The IR spectrum showed strong absorptions for hydroxy (3311 cm−1) and carbonyl (1671 cm−1) groups. The 1H NMR spectrum was initially obtained in CD3OD, but it had significant signal overlap in the aromatic region δH 6.00–7.50. The 1H NMR data in pyridine-d5 (Table 1) were more dispersed, and they showed signals for four methylene groups (δH 2.64, 3.12, 3.46 (2H), 3.67, 3.89, 4.17, 4.26) and five pairs of 2H doublets resulting from five para-substituted benzene groups (δH 6.79, 7.03, 7.09, 7.13, 7.15, 7.27, 7.31, 7.35, 7.41, 7.73). The combined 13C NMR and HSQC data (Table 2) confirmed the presence of 38 carbons including one carbonyl (δC 181.7), 12 nonprotonated sp2 carbons (δC 123.4, 126.4, 128.0, 128.5, 130.3, 132.6, 138.3, 158.1, 158.25, 158.33, 158.5, 158.8), 10 degenerate sp2 methines (δC 116.3, 116.6, 116.8, 117.2, 117.4, 129.3, 130.2, 130.3, 131.1, 132.8) that each reflected two aromatic carbons, one nonprotonated sp3 carbon (δC 61.8), and four methylene carbons (δC 31.3, 35.3, 38.8, 44.1). The five para-phenol groups that constitute rings A–E were deduced based on characteristic 13C chemical shifts, COSY couplings, and HMBC correlations (Figure 1). The deshielded chemical shifts of H-6 (δH 3.46, 3.89) and C-6 (δC 44.1), a COSY correlation from H-6 to H-7, and an HMBC correlation of H-9/C-7 suggested the A ring was part of a tyramine moiety. HBMC correlations of H-6/C-2 and H-6/C-5 indicated three C–N bonds linking C-2, C-5, and C-6 with N-1. HMBC correlations between H-16/C-14 and H-14/C-15 and C-16 helped establish a para-hydroxybenzyl group, which was attached at C-5 based on HMBC correlations between H-14/C-5. A 1H–15N HMBC of H-14/N-1 (δN 148.0) provided further support for this assignment. H-14 showed an additional correlation in the HMBC spectrum with C-4, revealing the connection between C-4 and C-5. An HMBC correlation between H-22/C-4 established the position of ring C. Ring E was part of another para-hydroxybenzyl substituent according to HMBC correlations of H-35/C-33 and H-33/C-34 and C-35. HMBC correlations of H-33/C-2, C-3, and C-4 helped define the linkage between C-2, C-3, and C-4 and the attachment of C-33 to C-3. Finally, ring D was linked at C-3 by an HMBC correlation of H-28/C-3. Although these assignments allowed the structure of 1 to be proposed, the positions of most aryl group substitutions around the central pyrrolone ring were based on a single HMBC correlation. Therefore, we conducted the recently reported LR-HSQMBC NMR experiment to detect additional four-bond and five-bond long-range C–H couplings.6 LR-HSQMBC correlations (optimized for nJC,H = 2 Hz) of H-14/C-21 and 27, H-22/C-5, H-28/C-2 and C-4, and H-35/C-3 provided further evidence for the position of the aryl substituents (Figure 1).
Table 1.
1H NMR Spectroscopic Data (600 MHz) for Compounds 1–7 [δH, mult. (J in Hz)]
| position | 1a | 2a | 3b | 4b | 5b | 6b | 7b |
|---|---|---|---|---|---|---|---|
| 5 | 4.52, t (4.4) | ||||||
| 6 | 3.46, mc | 4.01, t (8.2) | 3.37, m | 3.52, td (12.9, 5.3) | 3.62, mc | 3.62, mc | 3.88, t (8.0) |
| 3.89, ddd (14.0, 9.5, 4.7) | 4.05, ddd (13.6, 8.3, 5.0) | 3.71, td (12.8, 5.0) | 3.85, ddd (13.6, 11.9, 5.2) | 3.79, ddd (13.7, 11.9, 4.9) | |||
| 7 | 2.64, ddd (13.5, 9.1, 4.7) | 2.79, t (8.2) | 2.76, ddd (13.2, 7.8, 5.0) | 2.89, td (12.5, 5.3) | 2.93, ddd (12.7, 12.5, 5.3) | 2.98, m | 2.47, t (8.0) |
| 3.12, m | 2.87, m | 2.98, m | 3.08, ddd (12.4, 12.3, 5.1) | 3.10, ddd (12.2, 12.0, 4.8) | |||
| 9/13 | 7.09, d (8.3) | 7.07, d (8.5) | 7.02, d (8.4) | 7.15, d (8.1) | 7.22, d (8.4) | 7.22, d (8.3) | 6.56, d (8.3) |
| 10/12 | 7.13, d (8.3) | 7.13, d (8.5) | 6.73, d (8.4) | 6.75, d (8.1) | 6.78, d (8.4) | 6.79, d (8.3) | 6.51, d (8.3) |
| 14 | 3.67, d (16.9) | 4.34, s | 2.60, dd (14.5, 4.4) | 2.98, d (14.0) | 3.51, d (14.1) | 3.51, d (14.3) | 7.53, s |
| 4.26, d (16.9) | 3.03, dd (14.5, 4.4) | 3.21, d (14.0) | 3.62, mc | 3.62, mc | |||
| 16/19 | 6.79, d (8.1) | 7.40, d (8.6) | 6.60, d (8.6) | 6.57, d (8.3) | 6.25, d (8.7) | 6.25, d (8.7) | 7.38, d (8.2) |
| 17/20 | 7.41, d (8.1) | 7.17, d (8.6) | 6.57, d (8.6) | 6.53, d (8.3) | 6.21, d (8.7) | 6.21, d (8.7) | 6.93, d (8.2) |
| 22 | 7.31, d (8.6) | 7.63, d (8.6) | 6.94, d (8.7) | 7.43, d (8.5) | 8.50, d (8.8) | 8.31, d (8.8) | 8.56, d (9.8) |
| 23 | 7.03, d (8.6) | 7.16, d (8.6) | 6.68, mc | 6.71, d (8.5) | 7.34, dd (8.8, 2.4) | 7.34, dd (8.8, 2.4) | 7.28, m |
| 25 | 7.03, d (8.6) | 7.16, d (8.6) | 6.68, mc | 6.71, d (8.5) | 8.02, d (2.4) | 8.03, d (2.4) | 8.02, s |
| 26 | 7.31, d (8.6) | 7.63, d (8.6) | 6.94, d (8.7) | 7.43, d (8.5) | |||
| 28 | 7.73, d (8.5) | 7.63, d (8.6) | 6.86, d (8.6) | 6.87, d (8.2) | |||
| 29 | 7.35, d (8.5) | 7.16, d (8.6) | 6.68, mc | 6.68, d (8.2) | 7.92, d (2.4) | 7.93, d (2.4) | 7.93, s |
| 31 | 7.35, d (8.5) | 7.16, d (8.6) | 6.68, mc | 6.68, d (8.2) | 7.12, dd (8.8, 2.4) | 7.14, dd (8.8, 2.4) | 7.21, m |
| 32 | 7.73, d (8.5) | 7.63, d (8.6) | 6.86, d (8.6) | 6.87, d (8.2) | 8.84, d (8.8) | 8.86, d (8.8) | 9.08, d (8.8) |
| 33 | 3.46, mc 4.17, d (12.6) | 4.34, s | |||||
| 35/39 | 7.27, d (8.2) | 7.40, d (8.6) | |||||
| 36/38 | 7.15, d (8.2) | 7.17, d (8.6) | |||||
| 5-OCH3 | 2.97, s |
Measured in pyridine-d5.
Measured in CD3OD.
Signals overlapped.
Table 2.
13C NMR Spectroscopic Data (150 MHz) for Compounds 1–7 (δC, Type)
| position | 1a | 2a | 3b | 4b | 5b,d | 6b,d | 7b,d |
|---|---|---|---|---|---|---|---|
| 2 | 181.7, C | 128.2, C | 173.6, C | 171.6, C | 170.2, C | 169.8, C | 171.5, C |
| 3 | 61.8, C | 123.3, C | 131.5c, C | 132.2, C | 123.2, C | 124.9, C | n/a |
| 4 | 123.4, C | 123.3, C | 153.7, C | 151.5, C | 142.9, C | 139.2, C | 133.8, C |
| 5 | 138.3, C | 128.2, C | 63.5, CH | 94.7, C | 93.3, C | 98.5, C | 136.7, C |
| 6 | 44.1, CH2 | 47.6, CH2 | 44.3, CH2 | 43.6, CH2 | 43.0, CH2 | 42.7, CH2 | 44.7, CH2 |
| 7 | 35.3, CH2 | 37.8, CH2 | 34.7, CH2 | 35.6, CH2 | 36.4, CH2 | 34.9, CH2 | 34.4, CH2 |
| 8 | 130.3, C | 130.1, C | 131.1, C | 131.6, C | 131.5, C | 131.3, C | 130.1, C |
| 9/13 | 131.1, CH | 131.0, CH | 130.8, CH | 130.8, CH | 130.8, CH | 130.9, CH | 130.7, CH |
| 10/12 | 116.8, CH | 116.8, CH | 116.4,c CH | 116.3, CH | 116.3, CH | 116.3, CH | 115.9, CH |
| 11 | 158.1, C | 158.1, C | 157.3,c C | 157.0, C | 156.7,c C | 157.0,c C | 156.4, C |
| 14 | 31.3, CH2 | 31.0, CH2 | 36.1, CH2 | 41.6, CH2 | 43.6, CH2 | 42.6, CH2 | 116.6, CH |
| 15 | 128.0, C | 132.1, C | 127.1, C | 126.8, C | 126.7, C | 126.0, C | 127.7, C |
| 16/20 | 130.3, CH | 131.0, CH | 131.5,c CH | 131.8, CH | 131.2, CH | 131.5, CH | 132.5, CH |
| 17/19 | 117.4, CH | 117.0,c CH | 115.5, CH | 115.5, CH | 115.4, CH | 115.4, CH | 116.1, CH |
| 18 | 158.33, C | 157.9, C | 157.3,c C | 157.3, C | 156.7,c C | 157.0,c C | 158.6, C |
| 21 | 126.4, C | 129.0, C | 124.4, C | 124.9, C | 121.4, C | 121.2, C | 120.8, C |
| 22 | 130.2, CH | 132.6, CH | 131.5,c CH | 132.6, CH | 128.7, CH | 127.8, CH | 128.2, CH |
| 23 | 116.6, CH | 116.5,c CH | 117.0, CH | 116.0, CH | 118.5, CH | 119.3, CH | 119.0, CH |
| 24 | 158.25, C | 157.4, C | 161.5, C | 159.7, C | 158.8, C | 159.2, C | 158.4, C |
| 25 | 116.6, CH | 116.5,c CH | 117.0, CH | 116.0, CH | 108.7, CH | 108.9, CH | 109.0, CH |
| 26 | 130.2, CH | 132.6, CH | 131.5,c CH | 132.6, CH | 135.3, C | 135.1, C | 135.6, C |
| 27 | 132.6, C | 129.0, C | 124.0, C | 124.2, C | 121.7, C | 121.6, C | 121.4, C |
| 28 | 129.3, CH | 132.6, CH | 131.8, CH | 131.8, CH | 133.6, C | 133.7, C | 133.3, C |
| 29 | 117.2, CH | 116.5,c CH | 116.2, CH | 116.2, CH | 107.9, CH | 107.9, CH | 108.0, CH |
| 30 | 158.8, C | 157.4, C | 158.5, C | 158.5, C | 157.8, C | 158.0, C | 158.1, C |
| 31 | 117.2, CH | 116.5,c CH | 116.2, CH | 116.2, CH | 118.3, CH | 118.7, CH | 118.8, CH |
| 32 | 129.3, CH | 132.6, CH | 131.8, CH | 131.8, CH | 127.2, CH | 127.2, CH | 127.5, CH |
| 33 | 38.8, CH2 | 31.0, CH2 | |||||
| 34 | 128.5, C | 132.1, C | |||||
| 35/39 | 132.8, CH | 131.0, CH | |||||
| 36/38 | 116.3, CH | 117.0,c CH | |||||
| 37 | 158.5, C | 157.9, C | |||||
| 5-OCH3 | 51.3, CH3 |
Measured in pyridine-d5.
Measured in CD3OD.
Signals overlapped or interchangeable.
13C assignments were based on 1H–13C HMBC and HSQC correlations. The δc values were referenced to CD3OD (δC 49.00).
Figure 1.
Key 2D correlations for denigrin D (1).
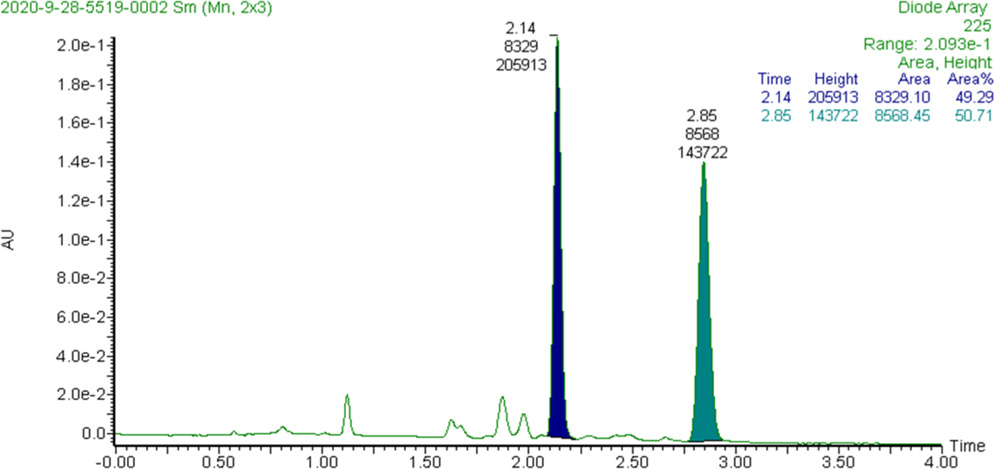
The denigrin D (1) sample did not show any Cotton effects in the ECD spectrum, but it did provide a measurable negative optical rotation (OR) at the sodium D wavelength, so assignment of the absolute configuration of C-3 was attempted. However, DFT-calculated OR values at five different wavelengths (365, 405, 436, 546, 589, and 633 nm) had poor correlations with the experimental OR measurements at these wavelengths, so no conclusions about the configuration of the stereogenic center could be drawn. Comparisons of the experimental and DFT-calculated VCD spectra were also inconclusive. The inability to closely correlate DFT-calculated physical properties such as OR and VCD with experimental measurements suggested that the sample might not be enantiomerically pure. So denigrin D was subjected to chiral-phase supercritical fluid chromatographic (SFC) screening across multiple chiral stationary phases,7 which revealed it was composed of a mixture of enantiomers (Figure 2). Chiral resolution of the enantiomers of denigrin D was achieved on a Chiralpak IC-3 chiral stationary phase, which established it as a racemic mixture based on peak area (≈1:1).
Figure 2.
Supercritical fluid chromatographic resolution of the enantiomers of denigrin D on a chiral stationary phase.
Denigrin E (2) was obtained as a magenta amorphous solid, and the molecular formula based on (+)-HRESIMS of C38H33NO5 had one less oxygen atom than 1. The IR spectrum showed a strong absorption band for hydroxy groups (3343 cm−1) and the absence of a carbonyl absorption. The 1H NMR (Table 1) and HSQC spectra of 2 showed only a limited number of signals including three signals for methylene protons (δH 2.79, 4.01, 4.34) and six aromatic signals (δH 7.07, 7.13, 7.16, 7.17, 7.40, 7.63). Careful integration of the 1H signals from most shielded to least shielded revealed a ratio of 1:1:2:1:1:2:2:2:2, respectively, suggesting a symmetrical structure for 2. COSY and HMBC correlations (Figure 3) confirmed a tyramine unit, two para-hydroxybenzyl units, and two para-hydroxyphenyl units attached to a pyrrole core. The HMBC correlation of H-16/C-5 (H-35/C-2) revealed the para-hydroxybenzyl groups were substituted at C-2 and C-5, while the correlation between H-22/C-4 (H-28/C-3) established the para-hydroxyphenyl groups at C-3 and C-4.
Figure 3.
Key 2D correlations for denigrins E–G (2–4).
Denigrin F (3) was purified as a dark yellow amorphous solid and gave a protonated molecule [M + H]+ in the (+)-HRESIMS spectrum at m/z 494.1952 that established a molecular formula of C31H27NO5 with 19 degrees of unsaturation. IR analyses confirmed the presence of hydroxy (3268 cm−1) and carbonyl (1655 cm−1) groups. NMR signals corresponding to eight aromatic doublets each integrating for two protons (δH/δC 6.57/115.5, 6.60/131.5, 6.68/116.2, 6.68/117.0, 6.73/116.4, 6.86/131.8, 6.94/131.5, and 7.02/130.8), one deshielded sp3 methine (δH/δC 4.52/63.5), three methylene groups (δH/δC 2.60 and 3.03/36.1, 2.76 and 2.87/34.7, 3.37 and 4.05/44.3), 10 nonprotonated sp2 carbons (δC 124.0, 124.4, 127.1, 131.1, 131.5, 153.7, 157.3 (2C), 158.5, 161.5), and a carbonyl carbon (δC 173.6) were observed in the 1H and 13C spectra (Tables 1 and 2). Following the 1D and 2D NMR analysis, an N,N-disubstituted tyramine moiety was established with the C-2 carbonyl and C-5 methine attached to the N atom, two additional para-hydroxyphenyl groups, and a para-hydroxybenzyl substituent (Figure 3). HMBC correlations from H-5 to C-2, C-3, and C-4 revealed the pyrrolone structure, while a key COSY correlation of H-5/H-14 and HMBC correlations of H-5/C-15, H-22/C-4, and H-28/C-3 defined the positions of the aryl substituents. Denigrin F (3) was optically active but showed no Cotton effects in the ECD spectrum. Its DFT-calculated OR values at five different wavelengths did not correlate well with experimental OR measurements, which suggested it also consists of a mixture of enantiomers.
Denigrin G (4) was isolated as an orange amorphous solid, and its molecular formula of C31H27NO6 was determined by (+)-HRESIMS measurements. The 1H and 13C NMR spectra of 4 (Tables 1 and 2) showed signals similar to those of 3, except for the loss of the N-substituted methine group in 3 and the appearance of a nonprotonated but oxygen-substituted carbon (δC 94.7) in 4. According to the NMR analyses and its molecular formula, denigrin G (4) was proposed to share the same structure with 3, but with a hydroxy substituent at the C-5 position (Figure 3). Denigrin G (4) is racemic since it was optically inactive and had no measurable Cotton effect in the ECD spectrum.
Dactylpyrrole A (5) was purified as a yellow amorphous solid, and its molecular formula was deduced to be C31H25NO6 from a protonated molecule [M + H]+ at m/z 508.1763 in the (+)-HRESIMS spectrum. A UV absorption maximum at 340 nm (MeOH) revealed extended conjugation in 5, and IR bands at 3311 and 1680 cm−1 suggested the presence of hydroxy and carbonyl groups. Unlike denigrins D–G (1–4), dactylpyrrole A (5) showed signals indicative of two ABX aromatic systems [δH 7.12 (dd, J = 8.8, 2.4 Hz), 7.34 (dd, J = 8.8, 2.4 Hz), 7.92 (d, J = 2.4 Hz), 8.02 (d, J = 2.4 Hz), 8.50 (d, J = 8.8 Hz), 8.84 (d, J = 8.8 Hz)] in the 1H NMR spectrum (Table 1), in addition to two para-substituted benzene groups [δH 6.21, 6.25, 6.78, 7.22 (2H each)] and three methylene groups (δH 2.93, 3.08, 3.51, 3.62 (2H), 3.85). The 13C NMR data of 5 (Table 2) were comparable to those of 4, suggesting significant similarities in their structures. HMBC correlations of H-6/C-2 and C-5, H-14/C-5, H-22/C-4, and H-32/C-3 supported the fact that 5 is related to 4 (Figure 4). Additional HMBC correlations from H-25 to C-28 and H-29 to C-26 revealed the connection between C-26 and C-28, thus forming a phenanthrene system composed of rings C–E. Dactylpyrrole A (5) was optically active but showed no Cotton effects in the ECD spectrum, and its experimental OR values did not correlate well with DFT-calculated ORs, so it is most likely a mixture of enantiomers.
Figure 4.
Key 2D correlations for dactylpyrroles A–C (5–7).
Dactylpyrrole B (6) was obtained as an amorphous yellow solid. The molecular formula of C32H27NO6 was established by (+)-HRESIMS with an [M + H]+ protonated molecule at m/z 522.1917. The 1H NMR spectrum (Table 1) was virtually identical to that of 5, except for the presence of a methoxy group at δH 2.97. The 13C data for 6 also corresponded closely with those of 5. The position of the methoxy group was assigned at C-5 according to its HMBC correlation to C-5 (Figure 4). Dactylpyrrole B (6) was optically inactive, and it may be an artifact that formed during fractionation of the extract via an acid-catalyzed reaction of compound 5 with methanol.
Dactylpyrrole C (7) was isolated as a yellow amorphous solid, and its molecular formula of C31H23NO5 was deduced by (+)-HRESIMS analysis. Dactylpyrrole C (7) was structurally related to 5 and 6 according to its 1H and 13C NMR data (Tables 1 and 2), but 7 lacked a hydroxy or a methoxy substituent at C-5 as well as the C-14 methylene. They were replaced by a nonprotonated olefinic carbon at δC 136.7 (C-5) and an sp2 methine at δH/δC 7.53/116.6 (C-14) in 7. This indicated the presence of a Δ5,14 double bond, which was further confirmed by HMBC correlations from H-14 to C-16 and C-4 and from H-6 to C-5 (Figure 4). Although the configuration of the double bond could not be directly defined due to the instability of 7, it was proposed as E based on the similar 1H and 13C NMR chemical shifts reported for the E olefin in denigrin B.8 A hypothetical aryl–aryl intramolecular coupling between rings C and D of denigrin B could lead to the connection between C-26 and C-28 and thus provide dactylpyrrole C (7).
The known compounds dictyodendrin F (8),9 denigrin A,8 denigrin B,8 and spirodactylone5 were also isolated from the Dactylia sp. extract, and they were assigned by spectroscopic data comparisons (NMR, MS, and specific rotations) with the literature values. Compounds 1–8, along with denigrins A and B, and spirodactylone were tested in a luciferase reporter assay developed to identify inhibitors of PAX3-FOXO1 translocation-induced gene transcription.10 Only dictyodendrin F (8) exhibited moderate, yet selective inhibition of PAX3-FOXO1-driven luciferase expression, with an IC50 value of 13 (±3) μM (Figure 5). It exhibited nonspecific cytotoxic activity at concentrations of ≥20 μM. Actinomycin D was the positive control in this assay with an IC50 of 1.0 nM. Other natural products that have shown activity in this type of PAX3-FOXO1 assay system include the cyclic tetrapeptide 1-alaninechlamydocin and thapsigargin, a highly functionalized sesquiterpene lactone.10 While the potency and selectivity of 8 were modest, these initial SAR results suggest that further exploration of aryl pyrroles as potential PAX3-FOX1 inhibitors should focus on elaboration of the substituted indole-2,6-dione core of dictyodendrin F.
Figure 5.
Dose–response curves for dictyodendrin F (8) in the PAX3-FOXO1 cell-based reporter assay. Compound 8 showed selective inhibition of PAX3-FOXO1 translocation-driven luciferase expression (ALK-Luc), relative to constitutively driven luciferase expression (CMV-Luc) in Rh4 cells.
EXPERIMENTAL SECTION
General Experimental Procedures.
Optical rotations were measured on a Perkin-Elmer 241 spectropolarimeter. UV spectra were measured with a Thermo Scientific Nanodrop 2000C spectrophotometer. IR spectra were recorded with a Bruker ALPHA II FT-IR spectrometer. NMR spectra were obtained with a Bruker Avance III NMR spectrometer equipped with a 3 mm cryogenic probe and operated at 600 MHz for 1H and 150 MHz for 13C. HMBC spectra were optimized for 8.3 Hz nJC,H and 8.0 Hz nJN,H and LR-HSQMBC for nJC,H = 2.0 Hz. The δN values were not calibrated to an external standard but were referenced to neat NH3 (δ 0.00) using the standard Bruker parameters. (+)HRESIMS data were acquired on an Agilent Technology 6530 Accurate-mass Q-TOF LC/MS. High-performance liquid chromatography (HPLC) was performed using a Varian ProStar 215 solvent delivery module equipped with a Varian ProStar 340 UV–vis detector, operating under Star 6.41 chromatography workstation software.
Animal Material.
Specimens of the Dactylia sp. nov. sponge were collected in the Republic of the Maldives in September 1997 and kept frozen until extraction. The collection was carried out by the Coral Reef Research Foundation under contract with the Natural Products Branch, U.S. National Cancer Institute. A voucher specimen (voucher ID # 0CDN5269) was deposited at the Smithsonian Institution, Washington, DC. The Dactylia sp. nov. sample had an encrusting, repent sponge body that was compressible and firm, with a smooth surface that contained oscules on the top of conules. The sponge skeleton presented a triangular to quadrangular network. It contained ascendant main fibers with connected fibers perpendicularly crossing them and secondary fibers forming circular meshes. Foreign material (fragments of spicules, sand grains, and foraminifers) were sparsely distributed, mainly in the nodes and connections of the fibers. This species of Dactylia is currently known only to the Maldives Islands, Indian Ocean (shallow waters). This represents a new species, which will be fully described in a later publication. The animal material (257.2 g, dry weight) was ground and processed using the standard NCI method for marine samples,11 to provide 5.43 g of organic solvent (CH2Cl2–MeOH, 1:1) extract (NSC # C017469).5
Isolation of Compounds.
As described previously,5 a portion of the organic extract (511.0 mg) was fractionated on Diol SPE cartridges (2 g) eluting with 9:1 (v/v) hexane–CH2Cl2, 20:1 (v/v) CH2Cl2–EtOAc, 100% EtOAc, 5:1 (v/v) EtOAc–MeOH, and 100% MeOH in a stepwise manner. The 100% EtOAc fraction was separated on a 50g C18 flash chromatography column, eluting with a linear gradient of MeOH–H2O (1:1–1:0, v/v) collected in four fractions (fractions A–D). Fraction A was subjected to reversed-phase C18 HPLC (Phenomenex Luna C18(2) 5 μ, 100 Å, 250 × 10.0 mm) using a linear gradient elution of MeOH–H2O [50:50–80:20, v/v, containing 0.1% trifluoroacetic acid (TFA)] over 30 min to afford dactylpyrrole A (5, 0.7 mg), denigrin B (2.0 mg), dactylpyrrole B (6, 0.3 mg), and dactylpyrrole C (7, 0.4 mg). Fraction B was purified by HPLC (Phenomenex Luna C18(2) 5 μ, 100 Å, 250 × 10.0 mm) with MeOH–H2O (30:70–70:30, v/v, containing 0.1% TFA) over 40 min to obtain denigrin D (1, 5.3 mg). Fraction C was dissolved in 2 mL of 50% MeOH and injected in 1 mL aliquots (~7 mg/injection) onto an HPLC column [Phenomenex Luna C18(2) 5 μ, 100 Å, 250 × 10.0 mm]. Solvent elution with MeOH–H2O (50:50–80:20, v/v, containing 0.1% TFA) over 30 min yielded seven subfractions (subfractions C1–C7). The two major UV-absorbing subfractions C5 and C6 were subjected to another HPLC using a Phenomenex Luna C18(2) (5 μ, 100 Å, 250 × 21.2 mm) column with gradient elution of MeCN–H2O (30:70–60:40, v/v). Subfraction C5 gave denigrin A (1.3 mg) and spirodactylone (0.4 mg), and subfraction C6 afforded denigrin F (3) and denigrin G (4) with less than 0.1 mg. Fraction D was purified by HPLC (Phenomenex Luna C18(2) 5 μ, 100 Å, 250 × 21.2 mm) to yield dictyodendrin F (8, 4.7 mg).
To afford a larger amount of compounds 3 and 4, a larger scale separation of the organic extract (1.5 g) was fractionated using the same procedure. A 50g C18 flash chromatography (same conditions as above) followed by an HPLC isolation [Phenomenex Luna C18(2) (5 μ, 100 Å, 250 × 21.2 mm), MeCN–H2O (30:70–60:40, v/v)] gave an additional 2.2 mg of 4, 1.4 mg of 3, and 7.4 mg of denigrin E (2).
Denigrin D (1): yellow amorphous solid; UV (MeOH) λmax (log ε) 228 (4.51), 279 (4.19) nm; IR (neat) νmax 3311, 1671, 1611, 1513, 1442, 1355, 1234, 1174 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 600.2395 [M + H]+ (calcd for C38H34NO6, 600.2381).
Denigrin E (2): magenta amorphous solid; UV (MeOH) λmax (log ε) 227 (4.27), 273 (3.95) nm; IR (neat) νmax 3343, 1611, 1512, 1445, 1359, 1236, 1172 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 584.2430 [M + H]+ (calcd for C38H34NO5, 584.2431).
Denigrin F (3): dark yellow amorphous solid; UV (MeOH) λmax (log ε) 229 (4.64), 277 (4.31), 313 (4.14) nm; IR (neat) νmax 3268, 1655, 1608, 1514, 1442, 1361, 1241, 1172 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 494.1952 [M + H]+ (calcd for C31H28NO5, 494.1962).
Denigrin G (4): orange amorphous solid; UV (MeOH) λmax (log ε) 227 (4.32), 275 (4.03), 324 (3.83) nm; IR (neat) νmax 3326, 1664, 1606, 1514, 1443, 1414, 1349, 1236, 1173 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 510.1912 [M + H]+ (calcd for C31H28NO6, 510.1911).
Dactylpyrrole A (5): yellow amorphous solid; UV (MeOH) λmax (log ε) 218 (4.04), 252 (4.10), 340 (3.48) nm; IR (neat) νmax 3311, 1680, 1610, 1516, 1445, 1209, 1142 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 508.1763 [M + H]+ (calcd for C31H26NO6, 508.1755).
Dactylpyrrole B (6): yellow amorphous solid; UV (MeOH) λmax (log ε) 224 (4.20), 251 (4.27), 341 (3.74) nm; IR (neat) νmax 3325, 1676, 1609, 1515, 1446, 1207, 1140 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 522.1917 [M + H]+ (calcd for C32H28NO6, 522.1911).
Dactylpyrrole C (7): yellow amorphous solid; UV (MeOH) λmax (log ε) 224 (4.17), 252 (4.20), 344 (3.72) nm; IR (neat) νmax 3300, 1680, 1609, 1515, 1445, 1209, 1141 cm−1; 1H NMR, Table 1; 13C NMR, Table 2; HRESIMS m/z 490.1661 [M + H]+ (calcd for C31H24NO5, 490.1649).
Chiral-Phase Analysis.
Chiral-phase SFC screening and optimization experiments were carried out on Waters Acquity UPC2 systems equipped with a fluid delivery module (a liquid CO2 pump and a modifier pump), a sampler manager–FL autosampler, two auxiliary column managers allowing six installed columns, a photodiode array detector, and MassLynx software. Columns packed with Chiralpak (AD-3, AS-3, IA, IB, IC, IE, IF) and Chiralcel (OD-3, OJ-3, OZ-3) were purchased from Chiral Technologies; Lux Amylose-2 and Lux Cellulose-4 columns were purchased from Phenomenex. Dimensions and particle size for all columns were 4.6 × 150 mm, 3 μm. Chiral SFC separations were carried out on the diverse set of columns described above by gradient elution at a flow rate of 3 mL/min with the back-pressure regulator set at 200 bar. The SFC eluents were solvent A (CO2) and solvent B (25 mM isobutylamine in MeOH). The mobile phases were programmed as follows: linear gradient from 1% to 40% B in 5 min; hold at 40% B for 1 min. The column and samples were maintained at a temperature of 40 and 20 °C, respectively.
PAX3-FOXO1 Assay.
Compounds were evaluated for their ability to inhibit PAX3-FOXO1-driven gene expression as described earlier.10 Briefly, compounds were tested against the PAX3-FOXO1 reporter cell line Rh4 ALK-Luc seeded at 3000 cells per well in 27 μL of medium in 384-well plates. Compounds were also tested in parallel against Rh4 CMV-Luc cells where luciferase reporter expression was placed under the control of a constitutively active promoter to gauge selectivity of compounds for inhibition of PAX3-FOXO1. After incubating cells overnight, they were treated with 3 μL of purified test compounds, chromatography fractions, or extract samples. After further incubation for 24 h, test plates were equilibrated to room temperature and treated with 30 μL of SteadyLite luciferase assay reagent, and luminescence signal was read using a plate reader. Luminescence signals from test wells were normalized as the percentage of those from corresponding DMSO treatment wells. Tests were performed in quadruplicate. Data were analyzed and IC50 values were calculated using GraphPad Prism.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Bokesch for assistance with HR-MS measurements, and W. Moser along with the collections staff of the Smithsonian NMNH, Department of Invertebrate Zoology. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jnatprod.0c01103.
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
Raw NMR data files (ZIP)
1D and 2D NMR spectra and HRESIMS for compounds 1–7 (PDF)
Contributor Information
Unwoo Kang, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States.
Laura K. Cartner, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Basic Science Program, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Frederick, Maryland 21702-1201, United States
Dongdong Wang, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States.
Chang-Kwon Kim, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States.
Cheryl L. Thomas, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Basic Science Program, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Frederick, Maryland 21702-1201, United States
Girma M. Woldemichael, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States; Basic Science Program, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Frederick, Maryland 21702-1201, United States
Berkley E. Gryder, Genetics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland 20892, United States
John F. Shern, Genetics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland 20892, United States
Javed Khan, Genetics Branch, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland 20892, United States.
Cristiana Castello-Branco, Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20560, United States.
Edward C. Sherer, Department of Analytical Research and Development, MRL, Merck & Co., Inc., Rahway, New Jersey 07065, United States
Xiao Wang, Department of Analytical Research and Development, MRL, Merck & Co., Inc., Rahway, New Jersey 07065, United States.
Erik L. Regalado, Department of Analytical Research and Development, MRL, Merck & Co., Inc., Rahway, New Jersey 07065, United States
Kirk R. Gustafson, Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702-1201, United States
REFERENCES
- (1).Linardic CM Cancer Lett. 2008, 270, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Nguyen TH; Barr FG Molecules 2018, 23, 2798/1–2798/22. [Google Scholar]
- (3).Van Soest RWM; Boury-Esnault N; Hooper JNA; Rützler K; de Voogd NJ; Alvarez B; Hajdu E; Pisera AB; Manconi R; Schönberg C; Klautau M; Picton B; Kelly M; Vacelet J; Dohrmann M; Díaz M-C; Cárdenas P; Carballo JL; Ríos P; Downey R World Porifera database. Dactylia Carter, 1885. In World Register of Marine Species; at http://www.marinespecies.org/aphia.php?p=taxdetails&id=166257, 2018. [Google Scholar]
- (4).Sato S; Kuramoto M; Ono N Tetrahedron Lett. 2006, 47, 7871–7873. [Google Scholar]
- (5).Kang U; Caldwell DR; Cartner LK; Wang D; Kim C-K; Tian X; Bokesch HR; Henrich CJ; Woldemichael GM; Schnermann MJ; Gustafson KR Org. Lett 2019, 21, 4750–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Williamson RT; Buevich AV; Martin GE; Parella TJ Org. Chem 2014, 79, 3887–3894. [DOI] [PubMed] [Google Scholar]
- (7).Barhate CL; Joyce LA; Makarov AA; Zawatzky K; Bernardoni F; Schafer WA; Armstrong DW; Welch CJ; Regalado EL Chem. Commun 2017, 53, 509–512. [DOI] [PubMed] [Google Scholar]
- (8).Kumar MMK; Naik JD; Satyavathi K; Ramana H; Varma PR; Nagasree KP; Smitha D; Rao DV Nat. Prod. Res 2014, 28, 888–894. [DOI] [PubMed] [Google Scholar]
- (9).Zhang H; Conte MM; Khalil Z; Huang X-C; Capon RJ RSC Adv. 2012, 2, 4209–4214. [Google Scholar]
- (10).Gryder BE; Wu L; Qi J; Woldemichael GM; Pomella S; Quinn TR; Park PMC; Cleveland A; Stanton BZ; Song Y; Rota R; Wiest O; Yohe ME; Shern JF; Qi J; Khan J Nat. Commun 2019, 10, 3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).McCloud TG Molecules 2010, 15, 4526–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.