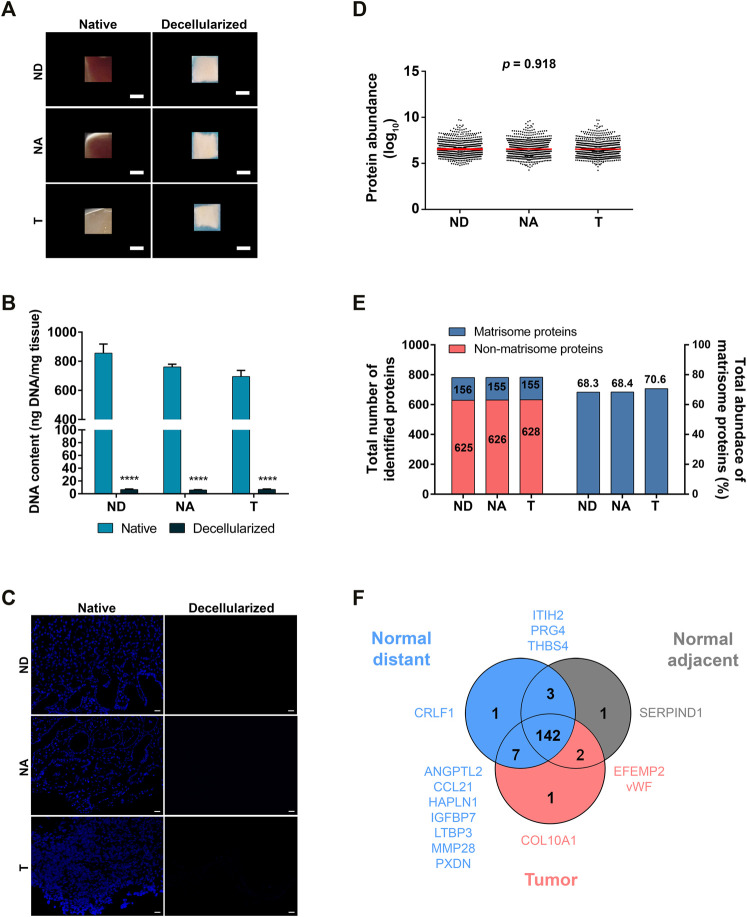
FIGURE 1.
Characterization and proteomic analysis of decellularized gastric ECM. (A) Representative macroscopic images of normal distant (ND), normal adjacent (NA) and tumor (T) samples from human gastric mucosa before and after decellularization; scale bar = 1 mm. (B) DNA quantification of native and decellularized ECM samples (n = 3). Data are shown as mean ± SD, ****p < 0.0001. (C) Representative images of DAPI staining of native and decellularized samples; scale bar = 20 µm. (D) Log10 of normalized abundance distribution for all identified proteins in decellularized samples. (E) Total number of matrisome and non-matrisome proteins identified in decellularized ECM, with the corresponding abundance (%) of each set of matrisome proteins. (F) Venn diagram with the number of matrisome proteins identified in each tissue in at least 1/3 of the samples (n = 9). We have identified a gastric matrisome signature composed of 153 proteins (in blue) and a tumor-specific ECM signature with three proteins (in red). Proteins present in only one or two ECM sets are specified.