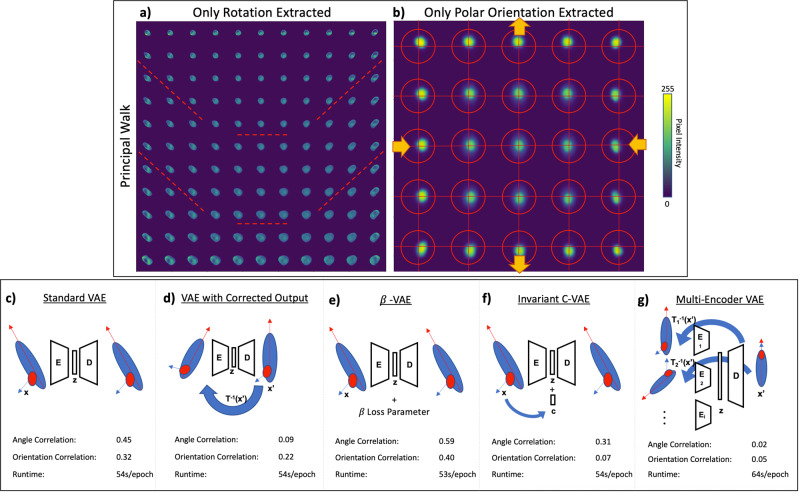
Fig. 1. VAE hypersensitivity and proposed model architecture.
VAE analysis of two datasets is shown, each governed by a single biologically uninformative feature a rotation and b polar orientation. Principal walk reconstructions23 show the VAEs’ governing features across the latent space through a range of image reconstructions. To correct this model hypersensitivity, several architectures were tested: c standard VAE with matched raw images; d VAE with paired randomly transformed input and controlled output images; e β-VAE that operates similarly to the Standard VAE, but utilizes a β hyperparameter in the loss function to encourage an independent latent space; f an invariant conditional VAE that injects the values of the uninformative features into the decoder such that they are not embedded in the latent space; g the proposed multi-encoder VAE: VAE with corrections for multiple features (rotation, polar orientation, size, shape, etc.) using parallel encoder models, a shared latent space, and a single decoder model. In c–e, a correlation between the embedding components and the respective feature (angle and orientation) is measured to quantify how effectively the model removes uninformative features. PBS and TGFβ + EGF cell populations with single channels were used in this analysis (n = 15,898 single-cell images).