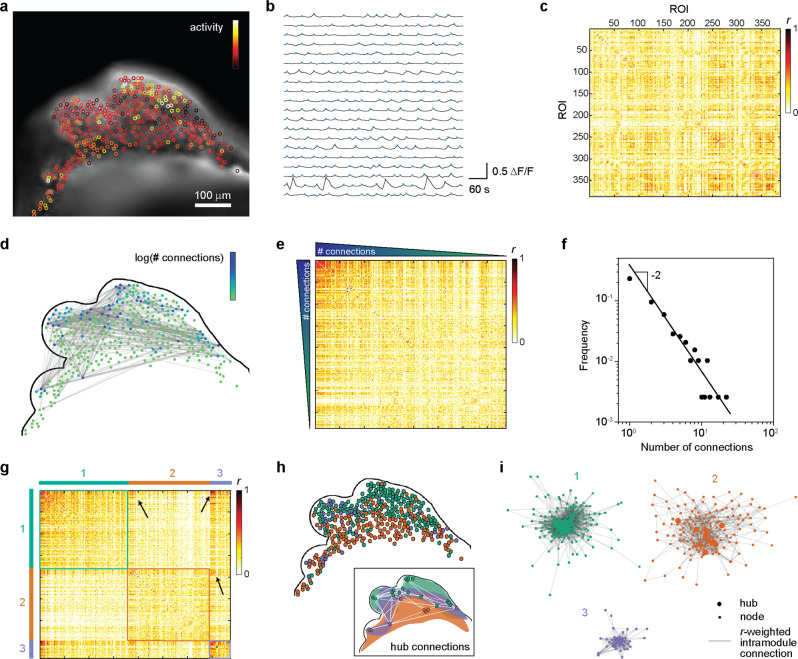
Fig. 2. Functional network interconnectivity in intact MARC-produced cerebral tissues.
a Neuronal activity in a cerebral tissue at week 4 of MARC culture. A snapshot of live fluorescence calcium imaging on the intact cerebral tissue is overlaid with regions-of-interest (ROIs) color-coded based on the frequency of detected calcium surges (“activity”) in each ROI. See Supplementary Movie 1 for the corresponding time-lapse imaging data. Scale bar: 100 µm. b Representative time traces of normalized intensity (ΔF/F) from 20 ROIs. Blue crosses indicate detected transient spikes. Time traces from 387 ROIs in the image are used to compute the correlation coefficient rij between ROI pairs i and j (see Methods for details of calculation). c Correlation matrix between any pair of the 387 ROIs, showing the r-value for each pair. d Depiction of functional connectivity network in the cerebral tissue. The contour of the cerebral tissue, corresponding to a, is shown. Two ROIs are defined to be functionally connected when r > 0.6 and shown as gray lines in the connectivity map. Further, each ROI in the connectivity map is color-coded based on the number of functional connections it has. e The same correlation matrix as in c, but with the ROIs sorted based on the number of functional connections they have. The high density of ROI pairs with a high number of connections and high r-value suggests a non-random network topology. f Distribution of the number of functional connections. The distribution follows a power law with a decaying power of −2, demonstrating a scale-free cerebral tissue functional network. g Clustered correlation matrix. To test whether the network exhibits modular topology, the functional connectivities are analyzed using the Louvain algorithm33, which indicates that the network contains three communities/modules. The correlation matrix in d is then reordered so that the nodes in the same module (color-coded with 1, 2, and 3) are positioned together. The high density of cross-module node pairs with high r-values (arrows) suggests the existence of hub connections between modules. h The localization of the nodes in the three modules. The color of the nodes corresponds to the color-coding of the three modules in g. The inset shows the spatial regions that enclose the nodes identified in the three modules, together with the hub nodes (defined as nodes with a number of intra-module connections larger than the 90th percentile in the module) and the cross-module hub connections (gray lines). i Topological representation of the intra-module functional connectivity networks. To illustrate the topological proximity of highly connected nodes, each module network is shown using the Fruchterman–Reingold algorithm44, where the length of the lines connecting nodes is proportional to 1 − r (i.e., short lines indicate a high correlation coefficient between the node pairs and the converse). The hubs in each module are also indicated. The central positioning of the hub nodes, as well as the close topological proximity between the hub nodes, highlight their status as intramodular connector nodes in the cerebral tissue functional network.