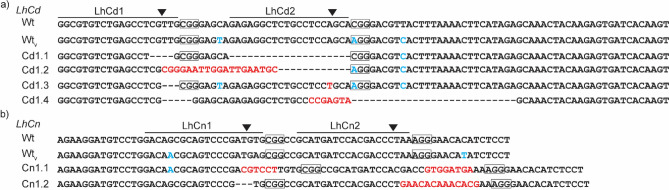
Figure 4.
Intergenerational LhCd and LhCn mutations in Lygus hesperus CRISPR/Cas9 mutant strains. Multiple sequence DNA alignments of intergenerational LhCd (a) and LhCn (b) mutations with the corresponding wild-type (Wt) sequences. Aligned regions of LhCd and LhCn only include sequences near the sgRNA target sites (LhCd1-2 and LhCn1-2). A variant allele within the wild-type sequences (Wtv) is shown. Horizontal lines indicate the sgRNA sequences, boxes indicate the PAM sequence trinucleotides, arrowheads indicate the predicted Cas9 cut sites, blue text indicates allele variants, red text denotes substitutions or insertions, and (–) corresponds to a sequence deletion.