Bedaquiline is a novel anti-tuberculosis drug for the treatment of multidrug-resistant tuberculosis (MDR-TB) recommended by the World Health Organization (WHO) [1] and recently upgraded to the group A classification of TB drugs as one of the three key drugs, along with linezolid and fluoroquinolones, to be included in all MDR-TB treatment regimens. Based on this grouping of second-line drugs, extensively drug-resistant tuberculosis (XDR-TB) is redefined as MDR- or rifampicin-resistant-TB that is resistant to a fluoroquinolone and to either bedaquiline or linezolid or both. Moreover, bedaquiline, in combination with pretomanid and linezolid, is a part of BPaL regimen recommended for treating adult pulmonary TB patients having pre-XDR-TB or MDR-TB which is either non-responsive or intolerant to recommended standard treatment [2]. However, globally emerging resistance to bedaquiline threatens the effectiveness of novel treatment regimens for drug-resistant TB.
Short abstract
This letter describes microevolution of a pre-XDR MTB strain isolated from a pulmonary TB patient over an 18-month exposure to BDQ. MDR-TB therapies with BDQ require a functional background regimen to prevent emergence of additional resistance. https://bit.ly/3D05qT9
To the Editor:
Bedaquiline is a novel anti-tuberculosis drug for the treatment of multidrug-resistant tuberculosis (MDR-TB) recommended by the World Health Organization (WHO) [1] and recently upgraded to the group A classification of TB drugs as one of the three key drugs, along with linezolid and fluoroquinolones, to be included in all MDR-TB treatment regimens. Based on this grouping of second-line drugs, extensively drug-resistant tuberculosis (XDR-TB) is redefined as MDR- or rifampicin-resistant-TB that is resistant to a fluoroquinolone and to either bedaquiline or linezolid or both. Moreover, bedaquiline, in combination with pretomanid and linezolid, is a part of BPaL regimen recommended for treating adult pulmonary TB patients having pre-XDR-TB or MDR-TB which is either non-responsive or intolerant to recommended standard treatment [2]. However, globally emerging resistance to bedaquiline threatens the effectiveness of novel treatment regimens for drug-resistant TB.
In this retrospective study, we discuss microevolution of a pre-XDR Mycobacterium tuberculosis strain isolated from a pulmonary TB patient at different time points over an 18-month period during treatment with a bedaquiline-containing regimen. We also report the acquisition of high-level resistance to bedaquiline and development of XDR TB due to emergence of an Ala63Pro mutation in the atpE gene encoding the bedaquiline target ATP synthase. Finally, we discuss the gradual disappearance of the Ala63Pro mutation followed by emergence and fixation of a premature stop codon mutation in Rv0678 during this period.
The first specimen for culture and drug susceptibility testing (DST) was collected before the start of treatment. DST was performed at a provincial laboratory by WHO-recommended rapid molecular DST (genotype MTBDRplus and MTBDRsl version 2; Hain Lifescience, Nehren, Germany) and pre-XDR-TB was reported. The diagnosis of pre-XDR-TB was subsequently confirmed by phenotypic DST with resistance to levofloxacin 1 mg·L−1 but susceptible to moxifloxacin 1 mg·L−1. Furthermore, pyrazinamide resistance was reported, along with sensitivity to second-line injectable drugs. The patient was initiated on a treatment regimen containing bedaquiline, moxifloxacin, linezolid, clofazimine, cycloserine, capreomycin, delamanid and ethionamide. The culture remained positive and after 9 months of treatment, failure was declared and treatment was stopped. After an interruption of 6 months, the patient presented at another treatment site and DST was repeated with similar results to those reported previously (pre-XDR-TB). The patient was re-enrolled on a treatment regimen containing bedaquiline, moxifloxacin, linezolid, cycloserine, ethionamide and pyrazinamide. The patient failed to culture-convert for 6 months; treatment was then modified and patient was continued on a bedaquiline, linezolid, clofazimine and delamanid containing regimen. Culture conversion was reported for the very first time, 8 months after re-enrolment and after 2 months on modified treatment, and thereafter remained negative till completion of 20 months of modified treatment. The patient was finally cured after treatment failure had been declared twice on bedaquiline-containing regimen.
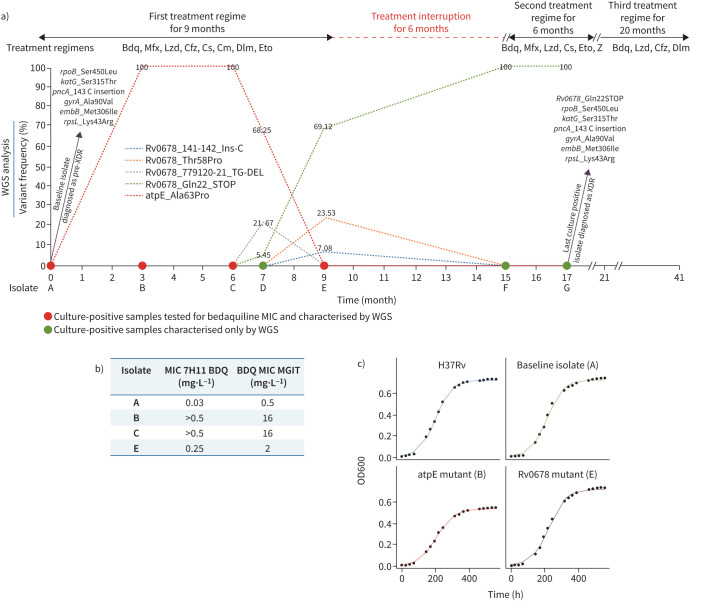
The isolates were collected by the national TB reference laboratory (NRL) as part of a surveillance project implemented to study acquisition of resistance to bedaquiline in Pakistan, as described previously [3]. Overall, seven Mycobacterium tuberculosis isolates from this patient were investigated, including a baseline (isolate A) and four cultures isolated on completion of 3, 6, 7 and 9 months (isolates B, C, D and E) of treatment and the last two isolates (F and G) at months 15 and 17 when the patient was re-enrolled (figure 1). At the NRL, all isolates were sub-cultured and DST was performed in a MGIT960 automated system (BD, Sparks, MD, USA) at WHO recommended critical concentration for bedaquiline, linezolid, clofazimine and delamanid [4]. Bedaquiline susceptibility testing for isolates F and G was also performed at 0.5 and 2.0, in addition to the critical concentration of 1.0 mg·L−1.
FIGURE 1.
a) Shown is a summary of genotypic drug resistance (based on whole genome sequencing (WGS)). b) Bedaquiline minimum inhibitory concentrations (MICs) performed in 7H11 and MGIT, and treatment monitoring during the bedaquiline-containing regimen. c) Growth curves of standing liquid cultures of Mycobacterium tuberculosis strains including H37Rv, baseline isolates (isolate A) and mutant strains (isolates B and E). The experiment was repeated three times giving comparable results and the mean values were plotted. Bdq: bedaquiline; Mfx: moxifloxacin; Lzd: linezolid; Cfz: clofazimine; Cs: cycloserine: Cm: capreomycin; Dlm: delamanid; Eto: ethionamide; Z: pyrazinamide; XDR: extensively drug resistant.
The strains were sent to the supranational reference laboratory (SRL), Milan in two shipments and included four live (A, B, C and E) and three inactivated culture isolates (D, F and G). Whole genome sequencing (WGS) was performed on all seven strains, with coverage depth threshold of 50× as described previously [3]. Moreover, all four live strains (A, B, C and E) were tested for the minimum inhibitory concentration (MIC) in 7H11 and MGIT as described previously [3]. Finally, for isolates A, B and E the analysis of growth rate through optical density measurements (OD600) on Middlebrook 7H9 medium (with OADC and 0.05% Tween 80) was undertaken as described elsewhere [5] and growth dynamics were plotted using the R package Growthcurver [6]. The H37Rv (ATCC 27294) strain was used as control strain in all experiments.
At SRL Milan, WGS of the baseline isolate revealed a Delhi-CAS sub-lineage strain with mutations conferring resistance to rifampicin, isoniazid, ethambutol, pyrazinamide, fluoroquinolones and streptomycin (figure 1a). This baseline isolate (A) was susceptible to bedaquiline having MICs of 0.03 and 0.5 mg·L−1 in 7H11 and MGIT, respectively (figure 1b). The WGS of the isolate collected at 3 months (B) after bedaquiline regimen was started unveiled acquisition of Ala63Pro mutation in atpE with variant frequency of 100%, which was not present in baseline isolate; this was associated with a very high bedaquiline MIC (>0.5 and 16 mg·L−1 in 7H11 and MGIT, respectively). Isolate C, collected after 6 months of bedaquiline exposure, showed the same bedaquiline MIC as isolate B and the persistence of this mutation in 100% of the bacterial population. The frequency of the atpE Ala63Pro decreased (variant frequency of 68.25%) in the subsequent isolate at month 7 (isolate D), but emergence of two different mutations in Rv0678 was seen (Gln22_STOP and a TG deletion at position 131–132 with variant frequencies of 5.45% and 21.67%, respectively). The Gln22_STOP mutant became the major population (variant frequency of 69.12%) in isolate E at 9 months of bedaquiline-containing treatment, but surprisingly the atpE_Ala63Pro mutation was not detected even by setting the variant frequency detection threshold of 1%. Moreover, the bedaquiline MIC of isolate E showed an eight-fold decrease with respect to isolates B and C (bedaquiline MIC 2 versus 16 mg·L−1 in MGIT), which was in accordance to disappearance of the atpE mutation. Furthermore, in isolate E the emergence of two other variants in Rv0678 were detected: a Thr58Pro substitution and a C insertion at position 141 with allele frequencies of 23.53% and 7.08%, respectively. Finally the Rv0678_Gln22_STOP mutation seems to become fixed (variant frequency of 100%) in isolates F and G collected, respectively, at 15 and 17 months from baseline, when the patient was restarted on a bedaquiline-containing regimen after 6 months of treatment interruption. These isolates were resistant at 1 mg·L−1 and sensitive at 2 mg·L−1 of bedaquiline in MGIT, confirming the MIC of 2 mg·L−1, the same as isolate E.
We report the transient emergence of a atpE_Ala63Pro mutation related to high-level bedaquiline resistance in a clinical Mycobacterium tuberculosis isolate. Moreover, our results show that multiple Rv0678 mutations may emerge independently and may be fixed during the treatment. However, the atpE_Ala63Pro mutant disappeared completely after 9 months of bedaquiline-containing regimen, which probably is related to the increased fitness cost of such a mutant. As a general rule for most bacterial species, resistance-conferring mutations can confer a biological cost that displays a selective growth disadvantage relative to the growth capability of drug-susceptible isogenic strains in the absence of the antibacterial drug [7]. In Mycobacterium tuberculosis, increased fitness costs have previously been shown for in vitro-selected rifampin-resistant mutants [8, 9], streptomycin-resistant mutants [10, 11], and isoniazid-resistant mutants [11, 12]. According to previous findings, the lower frequency of atpE mutations in clinical isolates potentially indicates a higher fitness cost of such mutants [13–15], whereas Rv0678 mutants of Mycobacterium tuberculosis seem to have the same fitness in comparison to the corresponding isolate, or even a little advantage [16]. Our results from calculation of generation time by plotting growth curves resulting from the OD600 at different time-points revealed that the atpE mutant has a slower growth rate and lower growth potential respect to the parental strain and the Rv0678 mutant, which confirms the lower fitness of atpE mutant in comparison to the parental strain and Rv0678 mutant (figure 1c). This finding is in line with previous studies, suggesting the concordance of growth rates measured in vitro with the frequencies at which these resistant mutants are recovered from patients in clinical settings [10, 11].
In conclusion, bedaquiline-containing regimens for MDR-TB require effective companion drugs to prevent the emergence of additional resistance and achieve high cure rates. Here we show that mutations affecting bedaquiline resistance behave differently due to their fitness cost. The emergence of target-based resistance within 3 month of bedaquiline treatment also shows the risk of resistance amplification in a relatively short time. Therefore, in countries with high burden of MDR-TB, it is crucial to develop capacity not only for phenotypic DST but also for MIC testing and WGS. Collection, storage and analysis of sequential isolates should be performed for patients who fail to culture convert within 3 months of treatment. TB programmes lacking capacity for phenotypic DST for new drugs and/or WGS need to secure resources and establish effective linkages with SRLs to temporarily support their WGS needs while developing in-country capacity.
Shareable PDF
Footnotes
This article has an editorial commentary: https://doi.org/10.1183/13993003.00149-2022
Raw sequencing reads for all isolates have been deposited at BioProject PRJNA750281 at NCBI.
Conflict of interest: None declared.
Support statement: This study was funded by a WHO project to NRL/NTP Pakistan and SRL Milan on the surveillance of acquisition of resistance to bedaquiline in Pakistan. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.World Health Organization . WHO Consolidated Guidelines on Drug-Resistant Tuberculosis Treatment. Geneva, World Health Organization, 2019. [PubMed] [Google Scholar]
- 2.World Health Organization . WHO Consolidated Guidelines on Tuberculosis. Geneva, World Health Organization, 2020. [Google Scholar]
- 3.Ghodousi A, Rizvi AH, Baloch AQ, et al. . Acquisition of cross-resistance to bedaquiline and clofazimine following treatment for tuberculosis in Pakistan. Antimicrob Agents Chemother 2019; 63: e00915-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization . Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines used in the Treatment of Drug-resistant Tuberculosis. Geneva, World Health Organization, 2018. [Google Scholar]
- 5.Peñuelas-Urquides K, Villarreal-Treviño L, Silva-Ramírez B,et al. . Measuring of Mycobacterium tuberculosis growth. A correlation of the optical measurements with colony forming units. Braz J Microbiol 2013; 44: 287–289. doi: 10.1590/S1517-83822013000100042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprouffske K, Wagner A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 2016; 17: 172. doi: 10.1186/s12859-016-1016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson DI. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol 2006; 9: 461–465. doi: 10.1016/j.mib.2006.07.002 [DOI] [PubMed] [Google Scholar]
- 8.Billington OJ, McHugh TD, Gillespie SH. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob Agents Chemother 1999; 43: 1866–1869. doi: 10.1128/AAC.43.8.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariam DH, Mengistu Y, Hoffner SE, et al. . Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2004; 48: 1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böttger EC, Springer B. Tuberculosis: drug resistance, fitness, and strategies for global control. Eur J Pediatr 2008; 167: 141–148. doi: 10.1007/s00431-007-0606-9 [DOI] [PubMed] [Google Scholar]
- 11.Böttger EC, Springer B, Pletschette M, et al. . Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat Med 1998; 4: 1343–1344. doi: 10.1038/3906 [DOI] [PubMed] [Google Scholar]
- 12.Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun 2002; 70: 4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nimmo C, Millard J, van Dorp L, et al. . Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 2020; 1: e165–e174. doi: 10.1016/S2666-5247(20)30031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimenkov DV, Nosova EY, Kulagina EV, et al. . Examination of bedaquiline- and linezolid-resistant Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother 2017; 72: 1901–1906. doi: 10.1093/jac/dkx094 [DOI] [PubMed] [Google Scholar]
- 15.Nieto Ramirez LM, Quintero Vargas K, Diaz G. Whole genome sequencing for the analysis of drug resistant strains of Mycobacterium tuberculosis: a systematic review for bedaquiline and delamanid. Antibiotics 2020; 9: 133. doi: 10.3390/antibiotics9030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Degiacomi G, Sammartino JC, Sinigiani V, et al. . In vitro study of bedaquiline resistance in Mycobacterium tuberculosis multi-drug resistant clinical isolates. Front Microbiol . 2020; 11: 559469. doi: 10.3389/fmicb.2020.559469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-02102-2021.Shareable (332.6KB, pdf)