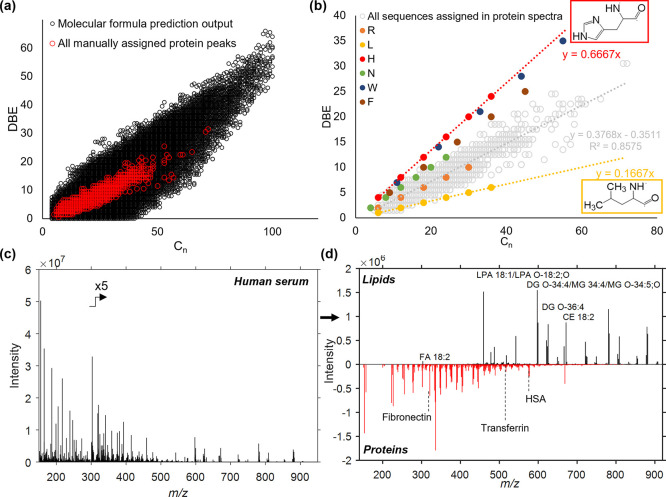
Figure 2.
(a) Comparison of all assignments proposed by MFP in a collated peak list of all manually assigned protein peaks in 16 different protein samples (black) and only correct protein assignments (red). (b) All protein peaks assigned in 16 protein samples are widely spread around the trend line of DBE/Cn (grey). The variability of DBE/Cn of protein assignments is caused by different amino acid DBE/Cn (orange—asparagine, R; yellow—leucine, L; red—histidine, H, green—asparagine, N; brown—phenylalanine, F). (c) Human serum sample is too complex to manually assign fragments of biological molecules. (d) MFP combined with the LIPID MAPS database automatically assigns lipid groups (black). Additional MFP on the rest of the spectrum enables assignment of proteins in serum (red). Peaks assigned as fragments of fibronectin (C15H22N4O3Na+, m/z 329.1585), transferrin (C24H44N6O5Na+, m/z 519.3258), and human serum albumin (C24H40N8O7Na+, m/z 575.2918). Chemical filtering was carried out on SIMS-MFP software.