Figure 1. Identification of novel cis-regulatory elements (REs) that modulate hTERT expression as a function of pluripotency.
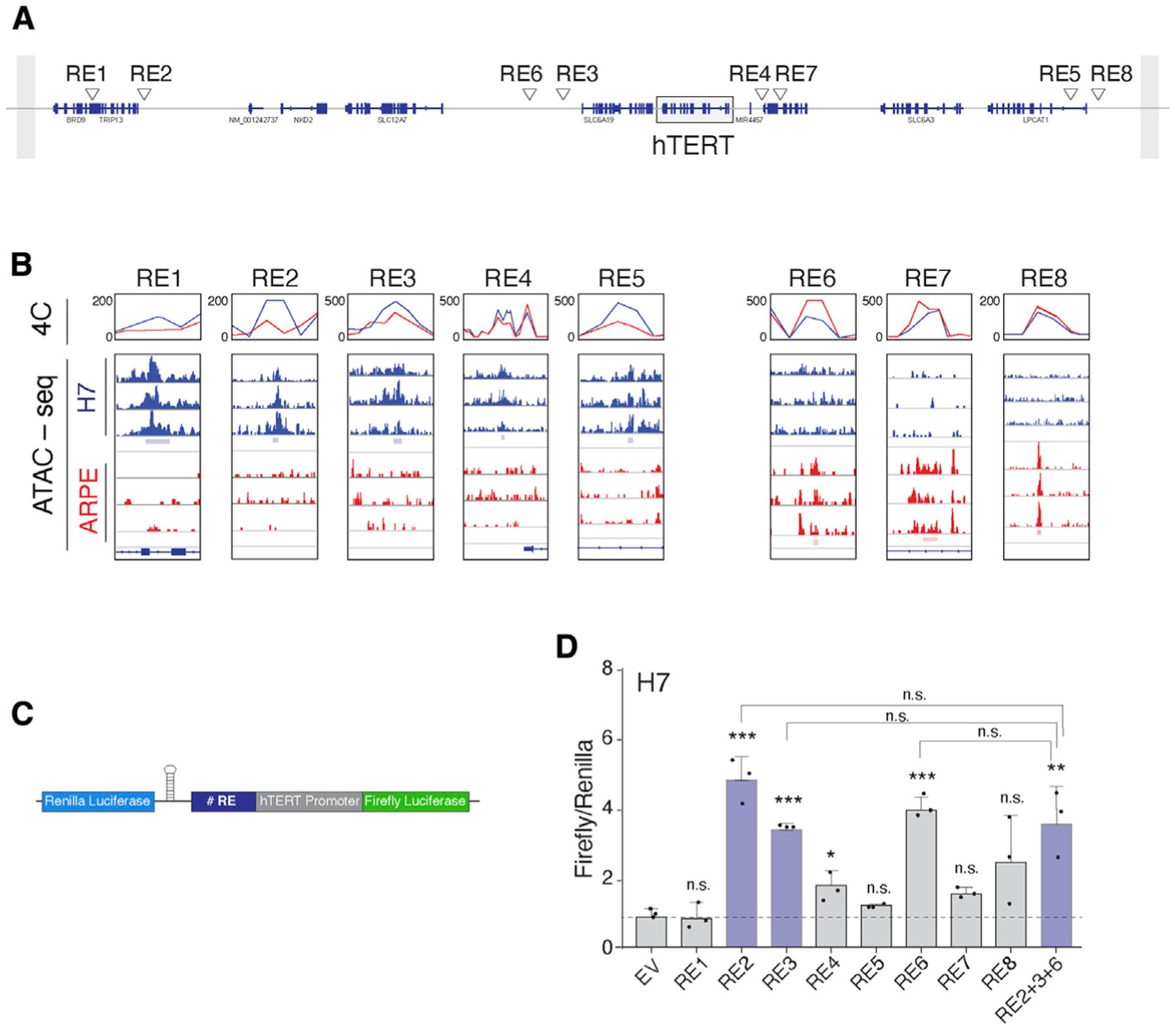
(A) Refseq of annotated genes on chr5, depicting candidate RE elements within the TAD boundary surrounding the hTERT locus (box).
(B) Overlay of data obtained from ATAC-seq and 4C-seq to identify candidate hTERT REs. Candidate enhancers (left, RE1–RE5) preferentially interact with hTERT promoter and display open chromatin in embryonic stem cells (ESCs). Silencer elements (right, RE6–RE8) preferentially interact with hTERT promoter in differentiated cells. 4Cker software was used to determine interaction score (line graphs) and aligned to ATAC-seq results from three independent replicates.
(C) Schematic of the dual-luciferase reporter used to interrogate the function of individual REs. hTERT core promoter: 500 bp sequence upstream of transcription start site (TSS) and 100 bp downstream of start codon. Individual REs cloned upstream of hTERT promoter. Expression of Renilla luciferase driven by a constitutive promoter for normalization. A hairpin downstream of the Renilla gene prevents transcriptional read-through.
(D) Bar graph represents the ratio of Firefly to Renilla luciferase in ESCs expressing the reporter plasmid with the indicated RE. n = 3 independent biological replicates. *p < 0.05, **p < 0.01, and ***p < 0.001, statistics calculated using ANOVA with multiple comparisons and comparing each value with empty vector (EV) control; similarly, ANOVA performed comparing RE2, RE3, and RE6 with RE2+3+6.
