Figure 4. Forced retention of exon 2 prevents silencing of hTERT upon differentiation.
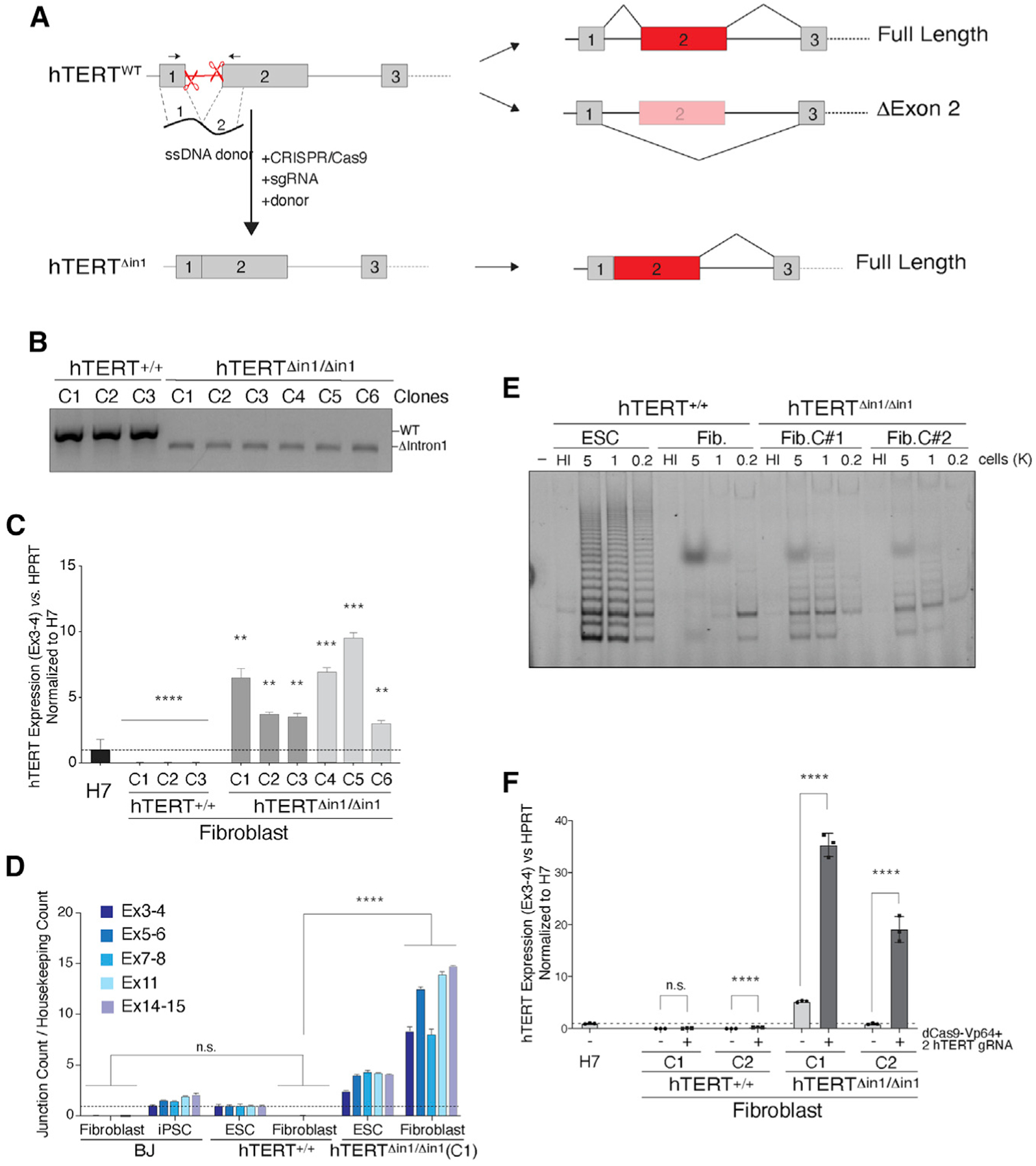
(A) Schematic illustration of intron 1 deletion by CRISPR-Cas9 gene editing and the predicted splicing pattern. Cells were co-transfected with two single guide RNAs (sgRNAs) that cleave within hTERT intron 1 and a 200 bp single-stranded (ss) DNA donor containing 100 bp sequence from exon 1 and exon 2 directly concatenated.
(B) Genotyping PCR from cells with the indicated genotype. PCR products: wild-type, 584 bp; Δintron1, 480 bp.
(C) qRT-PCR for hTERT mRNA in cells with the indicated genotype. Dark gray and light gray bars represent two independent CRISPR-Cas9 targeting experiments, and three independently derived clonal cell lines were generated from each. Values are normalized to hTERT+/+ H7 ESC. n = 3 biological replicates. **p < 0.01, ***p < 0.001, and ****p < 0.0001; statistics were computed using ANOVA with multiple comparisons, comparing hTERT expression with that of H7 ESCs.
(D) Absolute quantification of multiple hTERT exon-exon junctions using direct NanoString quantification. Data normalized to hTERT+/+ ESCs. n = 3 biological replicates. p < 0.001; statistics computed using ANOVA.
(E) TRAP assay for telomerase activity in cells with the indicated genotype shows that hTERTΔin1/ Δin1 fibroblasts retain telomerase activity compared with wild-type control cells. HI, heat inactivated.
(F) qRT-PCR for hTERT mRNA in differentiated fibroblasts following Vp64-Cas9 transcriptional activation of hTERT. Two hTERT+/+ and hTERTΔin1/Δin1 clones were transduced with lenti-virus particles expressing Vp64-dCas9 and two guide RNAs (gRNAs) targeting the hTERT promoter. Following fluorescence-activated cell sorting (FACS) selection of Vp64-dCas9-expressing cells, qRT-PCR was performed on non-transduced parental clonal lines and Vp64-dCas9-expressing cells. Values normalized to H7 cells. n = 3 biological replicates. p < 0.001, Student’s t test.
