Figure 6. Patient-derived SON mutation is associated with short telomeres and telomerase insufficiency.
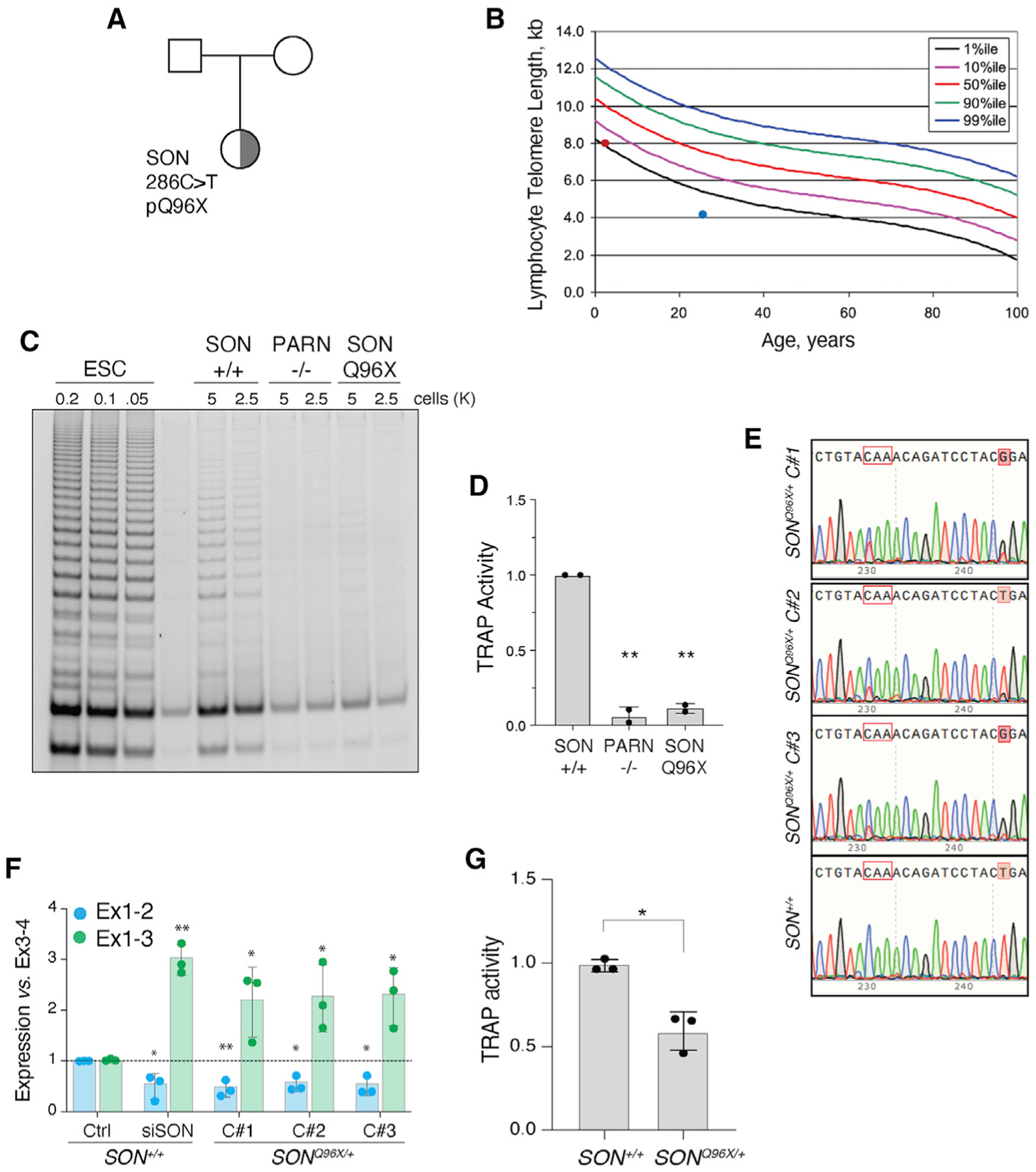
(A) Pedigree highlighting the proband, a female child of unaffected parents, carrying a de novo SON Q96X heterozygous mutation identified by exome sequencing.
(B) Telomere length measure by flow cytometry for total lymphocytes. Percentile curves are derived from healthy donors. Red dot represents lymph telomere length of SON carrier (NCI-550-1 with 8.1 kb telomeres). Blue dot represents a patient with PARN p.N7H mutation (NCI-382-1). Patient with lymph telomeres ~4.1 kb in length.
(C) Representative TRAP assay for telomerase activity in PBMC from a healthy donor (WT) and the proband (SON). PBMC from a PARN patient as a control.
(D) Quantification of the TRAP assay in (C) (p < 0.01). WT (TA 4646 0523), PARN p.N7H (c.19A > C) and deletion chr16:14,037,911 – 15,319,123 (NCI-382-1 – TA 2812 0860; patient published in Moon et al., 2015), SON mutation c.286C > T exon 3 p.Q96X (NCI-550-1 – TA 5330 0534).
(E) Sanger sequencing of three SONQ96X/+ independently derived clones and one SON+/+ clone. Tracks aligned to reference genome sequence. Codon in red outline encodes Q96. C > T mutation affects first nucleotide in codon. Shaded red boxes indicates an engineered silent T > G mutation to disturb gRNA binding site upon successful allele targeting.
(F) Quantification of the abundance of hTERT exon1-2 and exon1-3 splice junctions in SONQ96X/+ and SON+/+ ESCs. n = 3 biological replicates. *p < 0.05 and **p < 0.01, statistics calculated using ANOVA.
(G) Quantification of the TRAP assay in Figure S7F (p < 0.05).
