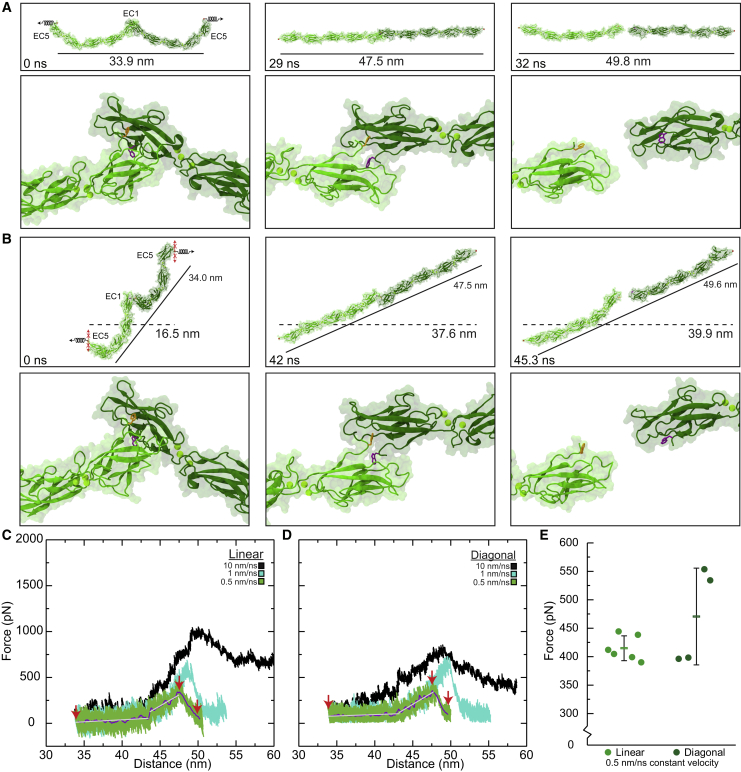
Figure 2.
Forced unbinding of trans CDH1 dimers in silico. (A) Snapshots of CDH1 unbinding at a stretching speed of 0.5 nm/ns (simulation S1d; Table 1). Stretched C-terminal Cα atoms are shown as red spheres. Springs in first panel indicate position and direction of forces applied along the vector joining the C-terminal Cα atoms (linear system) of the two monomers. The time and end-to-end distance between the C-terminal atoms are indicated in the top panels. Lower panels show the loss of the Trp2 exchange between CDH1 protomers. Trp2 residues are shown as orange and purple sticks for each monomer. (B) Snapshots of CDH1 unbinding at a stretching speed of 0.5 nm/ns (simulation S2d) shown as in (A). Force was applied with constraints (red Xs) to prevent motion perpendicular to the stretching direction (diagonal system). This mimics attachment to the cytoskeleton. All simulations showed straightening before unbinding without unfolding. Solid lines indicate end-to-end distance (between C-terminal Cα atoms) and dashes lines indicate membrane-to-membrane separation. (C) Force vs. end-to-end distance plot for linear constant velocity stretching of the CDH1 dimer at 10 nm/ns (S1b, black), 1 nm/ns (S1c, cyan), and 0.5 nm/ns (S1d, green; 1 ns running average shown in purple; gray lines are linear fits used to determine elasticity). Red arrowheads indicate time points in (A). (D) Force vs. end-to-distance plot for the diagonal constant velocity stretching of the CDH1 dimer shown as in (C) for simulations S2b-d. Red arrowheads indicate time points in (B). Forces in (C) and (D) are shown as monitored for one of the monomers. (E) Average magnitude peak force for simulations S1d-f (linear) and S2d-e (diagonal). Dots are force peaks from individual monomers. The bar represents the average from all protomers within a system (error bars are standard deviations).