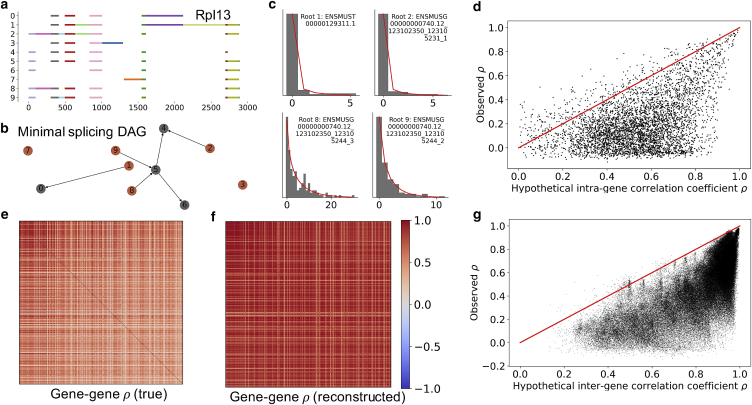
Figure 1.
Leveraging the synchronized burst model to predict transcript-transcript correlations. (a) By inspecting exon coexpression structures in long-read sequencing data, we can split genes into elementary intervals. (b) Although sequencing data are not sufficient to identify the relationships between various transcripts, they can provide information about “roots” of the splicing graph (highlighted in orange), which must be produced from the parent transcript by mutually exclusive processes. (c) The root transcript copy-number distributions are well described by negative binomial laws (gray histograms, raw marginal count data; red lines, fits). (d) The intrinsic noise model is not sufficient to accurately predict transcript-transcript correlations, but does serve as a nontrivial upper bound: few sample correlations exceed the model-based predictions obtained from Eq. 31 (points, transcript-transcript correlation matrix entries for mutually exclusive “root” transcripts of a single gene; red line, theory/experiment identity line). (e) The highest-expressed transcripts across the top 500 genes show distinctive, and generally positive correlation patterns. (f) We can use an analogous model to predict and reconstruct the correlation matrix based solely on marginal data. (g) As before, the model (Eq. 31) is not sufficient to accurately predict gene-gene correlations, but provides an effective and nontrivial upper bound (points, transcript-transcript correlation matrix entries; red line, theory/experiment identity line). To see this figure in color, go online.