Fig. 1.
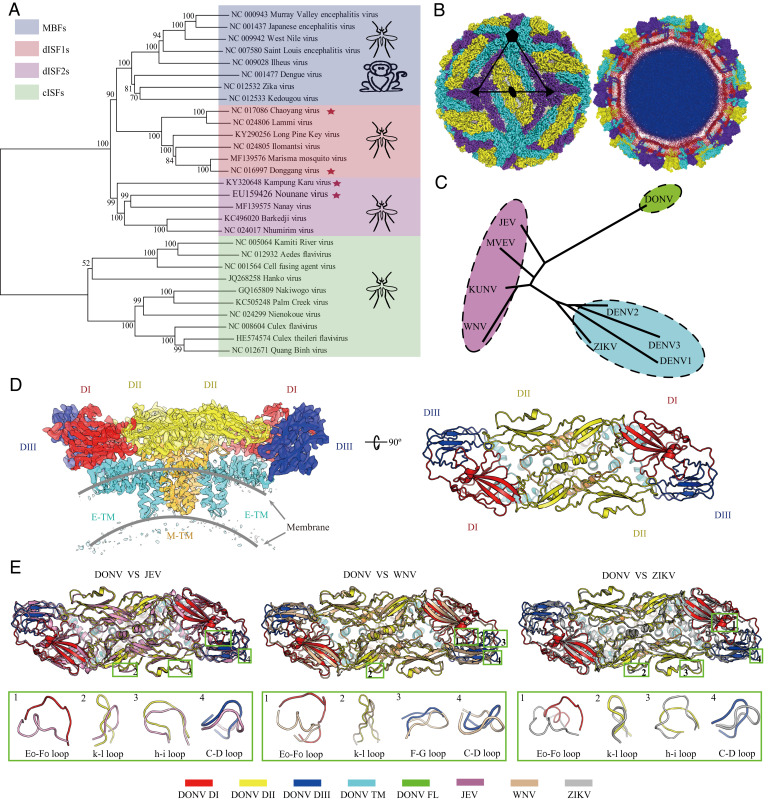
dISFs are phylogenetically and structurally close to MBFs. (A) Phylogenetic tree of mosquito-borne flaviviruses. Complete polyprotein amino acid sequences were aligned and a maximum likelihood phylogenetic tree was reconstructed in MEGA7. The consensus tree inferred based on 500 replicates is taken to represent the evolutionary history of the taxa. Asterisks indicate dISFs tested in Fig. 2. (B) 3D reconstruction (Left) and a thin slice of the central section (Right) of DONV viewed down an icosahedral twofold axis. E:M heterodimers of the same color are related by icosahedral symmetry. Heterodimers of different colors are quasiequivalent, with cyan E:M dimers falling on the icosahedral fivefold axes, purple on the threefold, and yellow on the twofold. (C) A structure-based phylogenetic tree constructed from E proteins of flaviviruses by Phylip-3.697: JEV (PDB code: 5WSN); KUNV, Kunjin virus (PDB code: 7KV9); MVEV, Murray Valley encephalitis virus (PDB code: 7KVB); WNV (PDB code: 3J0B); ZIKV (PDB code: 6CO8); DENV 1, Dengue virus 1 (PDB code: 4CCT); DENV 2 (PDB code: 7KV8); DENV 3 (PDB code: 3J6S); and DONV (PDB code: 7ESD). (D) Side view of the atomic model of the E:M:M:E heterotetramer shown in the ribbon. Domains I, II, III, transmembrane (TM) of E and M are colored in red, yellow, blue, cyan, and orange, respectively. (E) E and M structure superposition of DONV, JEV, WNV, and ZIKV. The structures of JEV, WNV, and ZIKV are shown in pink, brown, and gray. The obvious differences between DONV and other flaviviruses in conformation are highlighted by the green boxes.