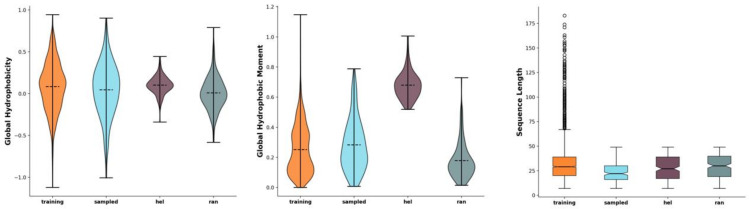
Figure 1.
Comparison of peptide characteristics between the training data (Training, orange), the generated sequences (Sampled, blue), the pseudo-random sequences with the same amino acid distribution as in the training set (Ran, purple), and the manually created hypothetical amphipathic helices (Hel, green). The horizontal dashed lines represent the mean (violin plots) and median (box plots) values; the whiskers extend to the outermost non-outlier data points. Graphs from left to right: Eisenberg hydrophobicity, Eisenberg hydrophobic moment, and sequence length. The figure was generated using the modlAMP’s GlobalAnalysis.plot_summary method in Python [16].