Figure 6. Sex-biased expression and trajectory analysis of testis cell lineages.
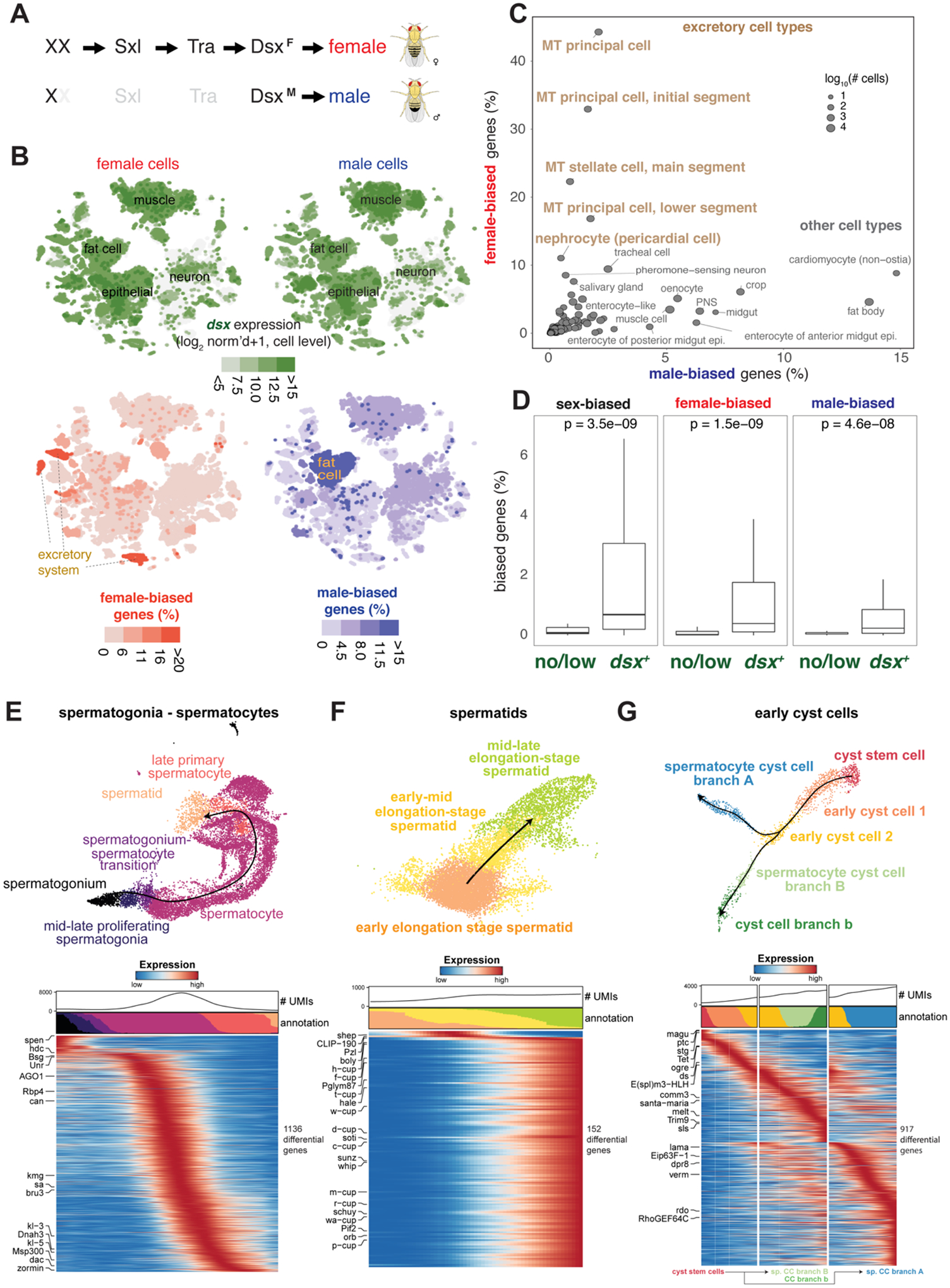
(A) Simplified sex determination pathway. Sex chromosome karyotype (XX) activates Sex-lethal (Sxl) which regulates transformer (Tra), resulting in a female Dsx isoform (DsxF). In XY (or X0) flies, Sxl and Tra are inactive (light gray) and the male-specific DsxM is produced.
(B) Top, Dsx expression and female- and male-biased expression projected onto tSNE plots of all female (left column) and male (right column) cells except reproductive tissue cells (Table S4 and S5). female- and male-biased expression measured as the percentage of genes in the cluster showing biased expression in favor of the respective sex (Table S6). These percentage values were computed for each annotated cluster and those cluster-level values were projected onto the individual cells in the corresponding clusters. For all four tSNE plots, values outside the scale in the heatmap key are represented by the closest extreme color (> and < signs in the scale).
(C) Scatter plot of female- and male-bias values across non-reproductive cell clusters defined as % sex-biased genes (at least 2-fold change with FDR < 0.05 on Wilcoxon test and BH correction) in the cluster (Table S6). Data point size indicatess cell numbers per cluster (key). Selected clusters are labeled, with those from excretory cells highlighted (brown). MT, Malpighian tubule.
(D) Box plots showing the relationship between dsx gene expression and sex-biased expression (Table S5). Clusters (B) were partitioned into the set of clusters with Dsx expression (dsx+) or not (no/low) using dsx expression in germ cells as an expression cut-off. Each box shows hinges at first and third quartiles and median in the middle. The upper whisker extends from the upper hinge to the largest value no further than 1.5 * IQR from the hinge (where IQR is the inter-quartile range, or distance between the first and third quartiles). The lower whisker extends from the hinge to the smallest value at most 1.5 * IQR of the hinge. Outliers are not shown. p-values are based on Wilcoxon test.
(E–G) Trajectory of testis subsets. We used slingshot to infer a possibly branching trajectory for spermatogonia-spermatocytes (E), spermatids (F), and early cyst cells (G). Shown are the trajectories on a UMAP (top) and the expression patterns of the strongest differentially expressed genes, together with the smoothed proportions of annotated cells and average number of unique molecular identifiers (UMIs) along the trajectory (bottom).
