Significance
Quantifying natural selection in human populations is a central topic in evolutionary biology and human genetics. Current studies to identify which single-nucleotide polymorphism has undergone selection suffer from limited sample sizes and large uncertainties in the timing of selection. In this study, we advance the field by showing that a genome-wide association study (GWAS) on infant mortality rate can identify recent selection signals. Our study produces well-powered genome-wide maps for selection. It replicates two selection signals that were detected in a previous study using ancient DNA, substantially improves the resolution on the timing of selection, and provides evidence for very recent selection during World War II. It also provides fundamental insights into how to interpret GWAS results.
Keywords: infant mortality, recent natural selection, regional GWAS
Abstract
Following more than a century of phenotypic measurement of natural selection processes, much recent work explores relationships between molecular genetic measurements and realized fitness in the next generation. We take an innovative approach to the study of contemporary selective pressure by examining which genetic variants are “sustained” in populations as mortality exposure increases. Specifically, we deploy a so-called “regional GWAS” (genome-wide association study) that links the infant mortality rate (IMR) by place and year in the United Kingdom with common genetic variants among birth cohorts in the UK Biobank. These cohorts (born between 1936 and 1970) saw a decline in IMR from above 65 to under 20 deaths per 1,000 live births, with substantial subnational variations and spikes alongside wartime exposures. Our results show several genome-wide significant loci, including LCT and TLR10/1/6, related to area-level cohort IMR exposure during gestation and infancy. Genetic correlations are found across multiple domains, including fertility, cognition, health behaviors, and health outcomes, suggesting an important role for cohort selection in modern populations.
A large literature over the past century has explored associations between phenotypic measures of health and social status and markers of fertility and reproductive success to draw inference about natural selection processes in the modern era (1). These studies highlight the still-evolving nature of human populations in low-mortality settings; they also facilitate prediction about short-run population change (2). Recent work provides new evidence on selection by leveraging large, biobank-scale genetic information, alongside new methods to summarize genome-wide measurements into polygenic scores (3) and tests of associations between summarized genetic information and reproductive success (4, 5). For example, studies have demonstrated associations between polygenic scores for specific phenotypes and reproductive success in the United States and in Iceland (6–8). Others have demonstrated single-nucleotide polymorphism (SNP) correlations with fertility markers through genome-wide association studies (GWAS) (9). Comparison of ancient genomes and modern human populations have implicated genetic loci for lactase persistence, skin pigmentation, immunity, and vitamin D metabolism (10). Still others have established genetic correlations between a broad set of phenotypes and fertility (11). For example, Sanjak et al. (11) finds genetic correlations between reproductive success and age at first birth, age at menarche, age at menopause, educational attainment (EA), and cognition, as well as body mass index (BMI) and related metabolic measures. In general, the findings are of modest directional selection that would accumulate into noticeable effects on larger time scales (8).
We advance this field by taking an innovative, complementary approach to the study of selective pressure. We ask which SNP alleles survive in populations when cohorts are exposed to better environments—or, in contrast, which alleles “disappear” in particularly harsh environments. We do this by using a “regional” GWAS (12). We measure the environment experienced by cohorts during gestation and in infancy, two periods in the life course prior to reproduction when cohort mortality is comparatively high (13, 14). We index the environment with the prevailing infant mortality rate (IMR) by place and year. The IMR is a well-established indicator of area-level conditions related to nutrition, infection, and inflammation, particularly during the early to mid twentieth century (15–17).
The approach builds on decades of demographic and epidemiologic research that considers how cohort traits are shaped by mortality exposure early in life (18–22). These studies provide several types of indirect evidence of early-life cohort selection: the differential survival of robust subpopulations through inhospitable disease environments during gestation and infancy [e.g., female versus male survival (23, 24)], nonlinear associations between disease environments and phenotypic traits like birth weight or height (25), and phenotypic traits among descendants of people exposed to war and famine in early life (26, 27). We shed light on these processes by explicitly examining molecular genetic information (SNPs) among cohorts who survive similar periods of hardship—and by testing how surviving SNPs are correlated with an array of complex traits, including the phenotypes explored in these previous studies.
We begin by examining associations between the early-life disease environment and the presence of specific loci among surviving cohort members. We then consider year-specific variation, including that aligned with particular hardships occurring during World War II. We conduct several sensitivity tests to rule out alternative explanations for the patterns detected in the UK Biobank (UKB) sample. We then examine the association between detected SNPs and the genetic predictors of an array of complex traits. We demonstrate that the shifts in genetic selection associated with improving early-life conditions are associated with genetic predictors of an array of reproductive, health, and behavioral traits.
Results
Infant Mortality Data.
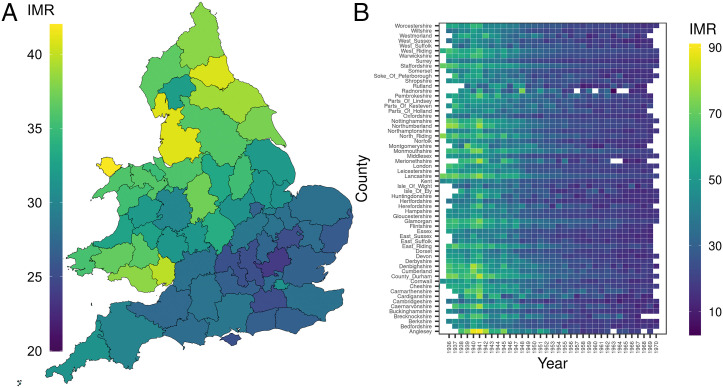
Area-level infant mortality fell over 60% over the time period of the UKB respondents’ birth years, between 1936 and 1970. Fig. 1 shows the time trend and the across-county variation in each year (SI Appendix, Fig. S1). These reductions were the result of improved living conditions and widespread public health efforts, among them, the expansion of prenatal and infant health care (28) after the war. Significant regional variation persisted through this period (29, 30). The IMR is highest in 1940 and 1941 when the UK population was exposed to intensive bombing campaigns during the World War II “Blitz,” alongside ongoing wartime reductions in nutrition that were particularly acute for unemployed and poor households (31, 32).
Fig. 1.
IMR between 1936 and 1970 in England and Wales. (A) Heatmap of average county-level IMR between 1936 and 1970 in England and Wales. IMR is defined as the percentage of deaths under 1 y of age (per 1,000 live births). The denominator of the IMR does not include stillbirths. (B) Heatmap of IMR in each county and year.
We measured IMR to capture the disease environment in the year prior to birth and during the year of birth in the county in which each participant in the UKB was born. We separately tested effects of these two exposures. The “lagged” IMR—or IMR during gestation—indexes conditions related to pregnancy survival in cohorts. The birth year IMR indexes conditions related to infant survival in cohorts. Throughout the study, we show findings from analysis of the birth year IMR. The findings of the analysis of gestation year IMR are presented in the SI Appendix.
GWAS Identifies Genetic Loci Associated with Birth Year IMR.
We conducted a GWAS on birth year IMR using 330,340 independent UKB respondents of European descent. To adjust for population stratification and the nonlinear time trend in IMR, we performed GWAS using the software BOLT-LMM (33) with year-of-birth effects included as indicator variables along with other covariates (Methods). Following previous work (12), we adjusted SE of SNP effects using the intercept from linkage disequilibrium (LD) score regression (34) to conservatively control type-I error.
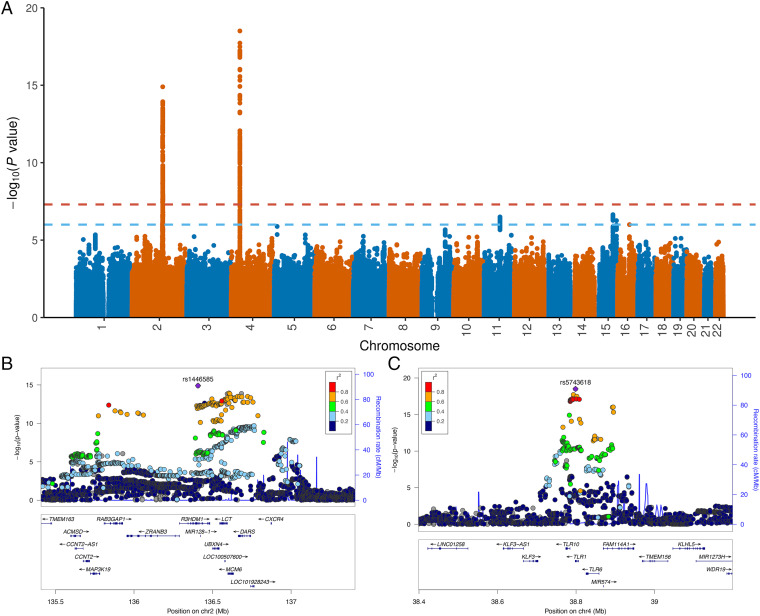
We identified two loci reaching genome-wide significance (Fig. 2 and Table 1): the lactase (LCT) locus on chromosome 2 (rs1446585; P = 1.26e-15) and the TLR1–TLR6–TLR10 gene cluster on chromosome 4 (rs5743618; P = 3.12e-19). Three loci showed suggestive associations (SI Appendix, Fig. S2): one locus near DHCR7 and NADSYN1 genes on chromosome 11 (rs2852853; P = 3.17e-7), one locus near the EFTUD1 gene (aka EFL1) on chromosome 15 (rs9944197; P = 2.27e-7), and one locus near RPGRIP1L and FTO genes on chromosome 16 (rs10521293; P = 9.99e-7). SNP heritability was low but statistically significant (h2 = 0.015, SE = 0.002), with an inflation factor = 1.06 (SI Appendix, Fig. S3). Heritability showed a twofold depletion in genomic regions annotated as heterochromatin but did not reach statistical significance after correcting for multiple testing (Dataset S1).
Fig. 2.
Genetic associations for birth year IMR. (A) Manhattan plot for birth year IMR. The horizontal lines mark the genome-wide significance cutoff of 5.0e-8 and a suggestive cutoff of 1.0e-6, respectively. (B) Genetic associations at the LCT locus. (C) Genetic associations at the TLR1/6/10 locus.
Table 1.
Genome-wide significant loci associated with birth year IMR
| CHR | SNP | BP (hg19) | A1 | A2 | EAF* | BETA | SE | P |
|---|---|---|---|---|---|---|---|---|
| 2 | rs1446585 | 136407479 | G | A | 0.232 | −0.157 | 0.020 | 1.26E-15 |
| 4 | rs5743618 | 38798648 | A | C | 0.233 | −0.176 | 0.020 | 3.12E-19 |
*Frequency of A1. Chromosome (CHR), variant ID, base-pair coordinate (BP) based on the genome reference build hg19, allele 1 (A1), allele 2 (A2), effect allele frequency (EAF), effect size (BETA), standard error (SE), and P value for the two genome-wide significant loci.
Conceptually, in order to focus on measures of the disease environment in the in utero period of each respondent, we used the IMR in the year prior to birth to capture the prevailing nutrition and infection conditions. We refer to this as the lagged-IMR phenotype. In practice, using the lagged or contemporaneous measure of IMR produces results with a genetic correlation of 1.001 (SE = 0.013; SI Appendix, Figs. S4 and S5). The main results reported in this paper are from the birth year IMR.
In order to guard against spurious findings, we also conducted falsification tests, where we performed a regional GWAS after randomly shuffling the county IDs and reassigning each participant’s IMR value (Methods). We found null results (SI Appendix, Fig. S6), which indicated that our findings were not driven by an artifact of the regional GWAS design.
GWAS Findings Suggest Recent Selection in the United Kingdom.
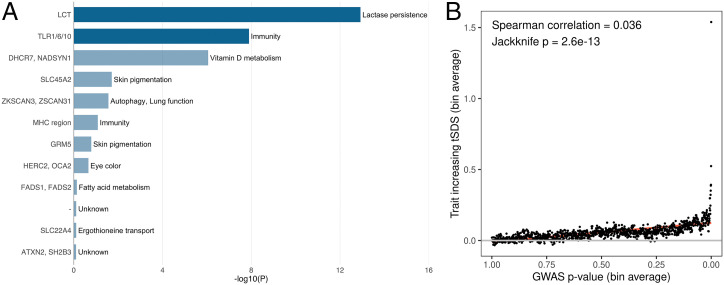
Our GWAS on birth year IMR identified two associated loci, that is, LCT and TLR1/6/10, both of which are well-known targets of selection in Europeans (10, 35–39). The beneficial alleles at both loci are associated with higher birth year IMR in our analyses, which is consistent with positive selection of these alleles in tougher environments. A recent study of more than 200 ancient genomes identified 12 target loci, including both LCT and TLR1/6/10, with strong signals of selection, possibly through their associations with nutrition, immunity, pigmentation, and other human traits (10). Among these 12 previously identified natural selection targets, genetic loci associated with lactase persistence (LCT), resistance to leprosy and other mycobacteria (TLR1/6/10), and vitamin D metabolism (DHCR7 and NADSYN1) showed strong associations with birth year IMR, whereas previously found loci for pigmentation did not show associations in our analysis (Fig. 3A and Dataset S2). Additionally, the strength of IMR GWAS associations is correlated with IMR-increasing single density scores (SDS; Spearman correlation = 0.036, P = 2.6e-13; Fig. 3B and Methods), suggesting polygenic and positive selection on IMR-associated alleles. We also confirmed this relationship using bivariate LD score regression (SI Appendix, Fig. S7; genetic correlation = 0.27; P = 0.0067). Notably, the LCT locus strongly colocalizes with high SDS, while the signal at TLR1/6/10 was not as strong (SI Appendix, Fig. S8).
Fig. 3.
Selection patterns at IMR-associated genomic loci. (A) Associations with birth year IMR at 12 target loci for selection in Europeans (10) (also see Dataset S2). Genetic loci reaching genome-wide significance in the IMR GWAS are highlighted in dark blue. (B) Correlation between GWAS associations of birth year IMR and SDS matched with IMR-associated alleles. The fitted linear regression line is shown in red.
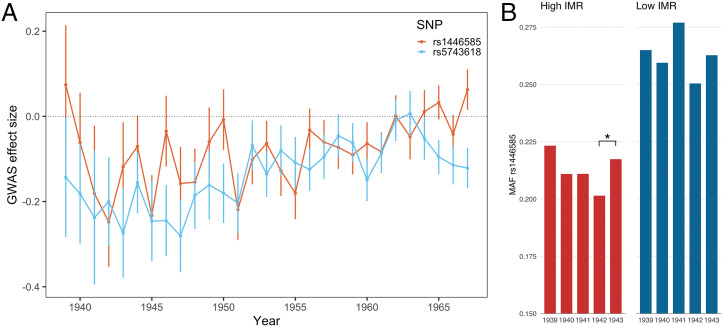
We further partitioned UKB samples by year of birth and estimated SNP–IMR associations for each year separately. Effect size at the LCT locus peaks in 1942, 1 y after “The Blitz,” and eventually bounces back to zero. The TLR1/6/10 locus shows a similar trend (Fig. 4A). In particular, the LCT locus (rs1446585) showed substantially attenuated associations with birth year IMR before 1941 and after 1942. We compared the allele frequency of rs1446585 in samples born in the top 10 counties with the highest IMR and in the 10 counties with the lowest IMR. We found substantial differences in allele frequencies between counties with high and low IMR. Interestingly, even within counties with high IMR, the lactase persistence–associated allele of rs1446585 (major allele A) showed increased frequency in the 1940 birth cohort, maintained similar frequencies through 1940–1941, further increase in frequency from 1941 to 1942, and then had significantly reduced frequency in samples born in 1943 (P = 0.018; Fig. 4B). The year-to-year comparison did not reach statistical significance in counties with low IMR (Dataset S3). We found null results for the TLR1/6/10 locus (SI Appendix, Fig. S9). We also note that the substantially elevated IMR in 1940 and 1941 is not correlated with the density and frequency of bombing events (SI Appendix, Fig. S10) and may be instead explained by other factors such as food scarcity during the war.
Fig. 4.
Selection on LCT and TLR1/6/10 loci during “The Blitz.” (A) The effect size of the lead SNP (rs1446585) at the LCT locus peaks in 1942 and eventually bounces back to zero. The lead SNP (rs5743618) at the TLR1/6/10 locus follows a similar trend. Dots and intervals indicate GWAS effect size estimates and SEs. Years with n < 5,000 were excluded from the analysis. (B) Minor allele frequency (MAF) of rs1446585 in UKB birth cohort in 1939–1943. Major allele at this locus is known to associate with lactase persistence. Counties with high/low IMR were defined based on IMR in 1939. The MAFs between 1942 and 1943 are statistically different at 0.05 level (*P = 0.018).
Genetic Correlation with 50 Complex Traits.
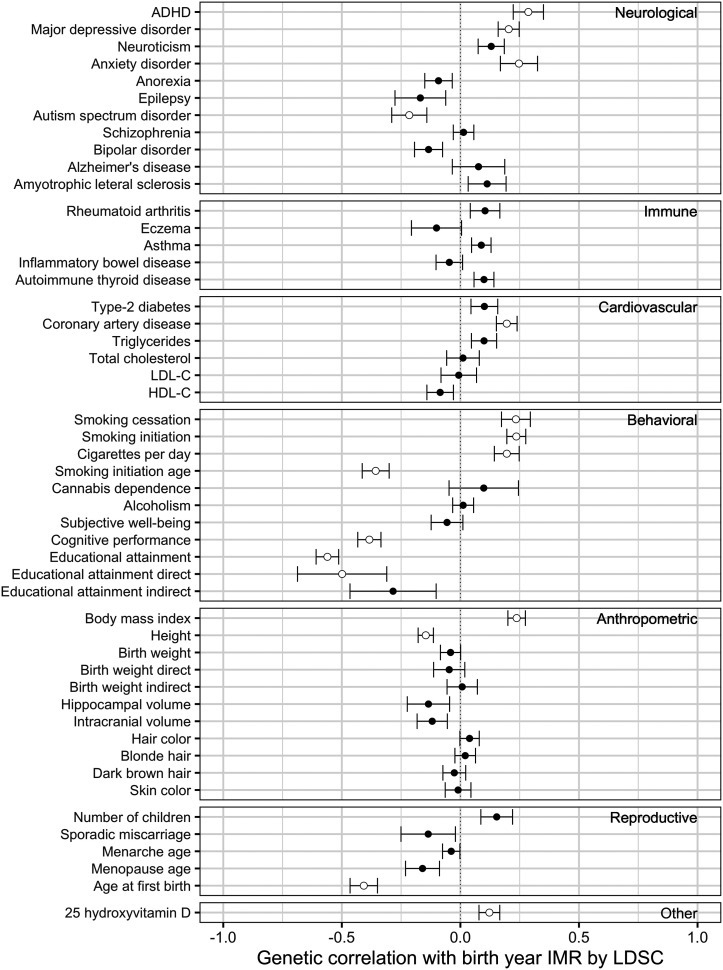
We next examined genetic correlation (40) between birth year IMR and a set of 50 traits widely assessed as outcomes of selection processes (Fig. 5 and Dataset S4). Among known target traits of selection (10), we found a significant correlation with vitamin D but found null results on hair and skin color. Our results are consistent with other approaches showing correlations with genetics of fertility (age at first birth) but do not find effects for number of children ever born, age at menarche, or age at menopause. Recall that Sanjak et al. (11) reported inconsistent findings between reproductive success and age at menarche (positive) and age at menopause (negative), which the authors label as “less explicable” than other results; as a comparison, we obtained null results for these two traits. Similar to earlier findings (6, 8), we show correlations with EA and cognition, but we extend this finding by showing these results are driven by the direct-EA component and not by the indirect-EA component mediated by family environment (i.e., genetic nurture), using methods in Wu et al. (41), suggesting that the selection pressure more directly applies to the child’s genetics on education rather than parental behavior that affects their children’s education. The difference in these findings suggest a broader need for caution when examining the genetic correlation findings, as we cannot decouple parental and child genetics in these results. We also find relationships with anthropometrics, like Sanjak et al. (11), but substantially extend our domains of interest to show findings for cardiovascular disease, tobacco use, and a variety of mental health conditions. The null findings on birth weight are suggestive that studies linking birthweight to insults akin to those prevailing during the 1930s and 1940s in the United Kingdom are likely capturing the deleterious effects of the disease environment (and accompanying wartime conditions) during that time, versus the differential survival of pregnancies (42, 43).
Fig. 5.
Genetic correlation between birth year IMR and 50 complex traits. Dots and intervals indicate genetic correlation estimates and SEs. Significant correlations at a false discovery rate cutoff of 0.05 are highlighted with circles. ADHD: attention-deficit/hyperactivity disorder; LDL-C and HDL-C: low (high) density lipoprotein cholesterol.
Discussion
In this work, we implemented an innovative approach to study natural selection in contemporary populations. We used GWAS to estimate how the frequency of common SNPs vary with area-level measures of infant mortality during the in utero period of UKB respondents and found two genome-wide significant signals at LCT and TLR1/6/10. These loci accord with previous work on natural selection comparing ancient and modern populations (10). We found limited evidence of large effects across the genome, and we estimated SNP heritability to be less than 2%. We then show moderate genetic correlations between the IMR GWAS and a host of phenotypic domains, reinforcing earlier findings related to fertility (age at first birth), anthropometrics (BMI, height), and cognition (EA, fluid intelligence), and also extending findings into psychiatric conditions (major depressive disorder, ADHD, anxiety disorder, autism) and health conditions (coronary artery disease).
Compared to past studies comparing allele frequencies in ancestral and modern populations (10, 44) and inferring lengths of the genealogy (36), this study directly estimates the shift of allele frequencies in less favorable environments. It identifies specific genomic loci under very recent selection and provides fundamental insights into the mechanism and timing of such selection. We found evidence for selection on lactase and potential resistance to leprosy and other mycobacteria in the past century and found null results for pigmentation traits. In particular, we demonstrate accelerated selection on these specific alleles in the United Kingdom during the mortality conditions caused by the World War II bombing campaigns during “The Blitz.”
We also note that, because our analysis is tied to area-level infant mortality, it differs from analysis that links reproductive success (number of children born) with genetic measurement (9, 11). In part, this is because the parents of the cohorts studied here who lose a pregnancy or an infant during periods of high infant mortality may have a subsequent successful pregnancy and surviving infant and thereby achieve levels of reproductive success. Instead, the focus here is on the genetic characteristics of people born between 1936 and 1970 who survive through reproductive age. We find that these survivors have genetic traits that are correlated with early initiation of childbearing. Like past research, we generally are unable to separate child (direct) and parental (indirect) genetic relationships (45) in the analysis. The results for EA suggest the effects are direct; that is, selection is occurring at the level of the child’s genetics rather than through family-level correlates of childbearing and survival, such as socioeconomic status.
Our study has a few limitations. First, although we provide evidence on the timing of selection and its ongoing nature, the specific mechanisms underlying such selection remain unclear. Even in the World War II example, we cannot parse out effects of stress, nutrition, or other related mechanisms. Although we demonstrate that these findings are unlikely due to participation bias, we cannot distinguish infant death from mortality later in life, although we note that infant mortality accounts for a large share (30 to 40%) of cohort mortality before age 55 y (average age of samples) in these birth cohorts. More generally, the study is not intended to pin down the effects of IMR specifically, but rather to describe how the cohorts’ mortality exposure, indexed here by IMR, is associated with the genetic composition of survivors at older ages.
Second, a limitation for most current research using genetic information is the focus on respondents of European ancestry. This study also faces this key limitation. Approximately 10% of the UKB sample was born in the United Kingdom but has African, Asian, or other ancestry. Data to support estimation of GWAS in populations with ancestry outside of Europe is growing and much needed. At present, the findings of this study are limited in applicability to the UK population with European ancestry.
Third, it remains a challenge to account for nonrandom participation of UKB (46, 47). We conducted several falsification tests to assess the impact of participation bias in our analysis. Adjusting for participation activities in GWAS and conditioning on a latent proxy factor for participation of optional UKB questionnaires (Methods) yielded highly consistent association results compared to our primary findings (SI Appendix, Figs. S11–S13). However, the latent propensity of participating additional questionnaires may be different from the propensity of participating UKB itself. Additionally, migration behavior in the UK is known to have a genetic basis (12), and the design of UKB will lead to undersampling in counties far from assessment centers, which makes migration another potential factor contributing to participation bias. We performed two additional GWAS using only 1) counties of birth with sufficient samples and 2) samples whose county of birth is identical to the county of current address (Methods). These GWAS revealed very similar associations compared to our initial analysis (SI Appendix, Fig. S14), with LCT and TLR1/6/10 being the genome-wide significant loci. Based on these results, we are cautiously optimistic that nonrandom sample selection in UKB may not severely bias our results. But, in general, participation bias remains a challenging open issue that remains to be investigated in the future.
Fourth, in order to guard against population confounding in our analysis, we employed the state-of-the-art linear mixed model approach (33) in conjunction with a conservative genomic control based on the software LDSC (48), and performed GWAS on a relatively homogeneous subpopulation in the UKB (49). However, unmeasured long-run characteristics at the county level (e.g., regional differences in socioeconomic status) may still partially confound genetic associations for IMR. To investigate this, we adjusted for household income, education years, and lagged IMR in GWAS (Methods) and obtained highly consistent association results (SI Appendix, Fig. S15), suggesting that long-run regional differences that can be captured by these covariates may not severely confound the IMR GWAS. Still, other county-level factors not included in our analysis remain to be investigated in the future. It is also important to note that, even if the approaches we have taken can effectively guard against false associations at the SNP level, it is not obvious whether minimal yet pervasive biases across the genome would lead to false findings in polygenic analyses such as genetic correlation (49, 50). It has been shown that bivariate LDSC provides robust estimates for genetic correlations, except when the population stratification biases in both input GWAS are correlated with LD scores (51). This strengthens our confidence in the genetic correlation results, since residual confounding in the IMR GWAS alone will not lead to biased genetic correlations as long as GWAS of other complex traits do not share a similar confounding. To further demonstrate this, we adjusted the IMR GWAS effect sizes by regressing out SDS which has been shown to have population stratification biases (49) (Methods), and we recomputed the genetic correlations with 50 complex traits. We obtained almost identical results compared to our primary analysis (SI Appendix, Fig. S16). Similarly, adjusting for socioeconomic status and lagged IMR also revealed consistent genetic correlations (SI Appendix, Fig. S17), suggesting limited evidence that our polygenic results are severely biased by unadjusted confounding.
Taken together, the research makes multiple contributions to the study of selective pressure using molecular genetic data. For example, several landmark studies (10, 36) have found specific genomic loci under selection over the time span of several millennia. The approach we use here, a regional GWAS, is designed to be exploratory and hypothesis-free, facilitating detection of previously undescribed relationships for contemporary cohorts. It is remarkable, then, that the approach in this study finds evidence of selection on the same loci as previous explorations using ancient genomes. In doing so, we demonstrate that selection described over the long arc of human history is occurring in the contemporary era. Further, we aggregated SNPs into domains of phenotypes through genetic correlation assessments and demonstrated that the shifts in genetic selection associated with improving early-life conditions are associated with an array of reproductive, health, and behavioral traits. These results shed important light on how selective pressure is correlated with complex traits in contemporary populations.
Methods
IMR Data in England and Wales.
We used a mortality table (“mort_lgd_ew”) produced by A Vision of Britain through Time (52) to obtain IMR and lagged IMR (i.e., IMR 1 y prior to the birth). A Vision of Britain through Time provides the year-specific number of births and deaths under age 1 y at the district level with the administrative county information from 1911 to 1974. We constructed a county–year-specific IMR from 1930 to 1970 by aggregating the number of births and deaths under age 1 y of all districts within each administrative county. Then the IMR was computed as 1,000 times the ratio of the number of deaths under age 1 y to the number of births for each county–year combination.
Using the publicly available boundary data for administrative counties as of 1931, 1951, 1961, and 1971 from A Vision of Britain, we classified the UKB participants in the administrative counties to cross-walk the county–year-specific IMR. Because the county–year-specific information on births and infant mortality was collected annually, while the boundary information was available only for the census years (i.e., 1931, 1951, 1961, and 1971), we classified UKB participants based on the year nearest to the (lagged) birth year, except for IMR in 1965 and lagged IMR in 1966 in which Greater London was established. Specifically, although the year nearest to the birth year of the 1965 cohort is 1961, we used the 1971 boundary data to classify the UKB respondents, since the data for 1961 did not have the boundary information for Greater London. Overall, to construct the IMR variable, we used the 1931 boundary data for those who were born between 1934 and 1941, the 1951 boundary data for those who were born between 1942 and 1956, the 1961 boundary data for those who were born between 1957 and 1964, and the 1971 boundary data for those who were born between 1965 and 1971. We constructed the lagged IMR in the same way. We dropped UKB participants born outside of England and Wales, because the mortality table produced by A Vision of Britain covered information only for England and Wales. Further, UKB participants who were born outside of the places the boundary data covered were also excluded from the analytical sample.
For visualization purposes, we downloaded county polygon shape files for England and Wales from A Vision of Britain through Time. We used the “sf” R package to process spatial data and plotted maps. We used the counties and boundaries for 1961 in Fig. 1. Middlesex and London became part of “Greater London,” Soke of Peterborough and Huntingdonshire became “Huntingdonshire and Peterborough,” and Cambridgeshire and Isle of Ely became “Cambridge and Isle of Ely” in 1964. In Fig. 1, we used the records from “Greater London” between 1965 and 1970 for Middlesex and London. Similarly, we “recovered” the 1965–1970 records for Soke of Peterborough, Huntingdonshire, Cambridgeshire, and Isle of Ely.
GWAS Analysis in the UKB.
Following Abdellaoui et al. (12), we deployed a regional GWAS in UKB to test associations between genetic measures at the individual-level and regional-level IMR, where all subjects reporting the same place of birth (data fields 129/130) and same year of birth (data field 33) had the same regional phenotypic value assigned. Of the 500,000 participants in UKB, we focused on the respondents with European ancestry defined as the principal component analysis inferred “Caucasian” participants (UKB data field 22006) which is a subset of self-reported “White British” individuals (data field 21000), and those with places of birth in England and Wales (data field 1647) in order to match with our contextual data. We excluded the participants who are recommended by UKB to be excluded from analysis (data field 22010), those with conflicting genetically inferred (data field 22001) and self-reported sex (data field 31), and those who withdrew from UKB. UKB samples with European ancestry were identified from principal component analysis (data field 22006) of the genotypes. We used software KING (53) to infer the pairwise family kinship among UKB samples and identified 154 pairs of monozygotic twins, 242 pairs of fraternal twins, 19,136 full sibling pairs, and 5,336 parent–offspring pairs among 408,921 individuals of European descent. A total of 330,340 independent samples from 65 unique counties who were born between 1934 and 1970 were used in the IMR GWAS. We used BOLT-LMM (33) to perform GWAS with sex, genotype array (data field 22000), and year of birth as covariates. Year of birth was dummy coded to account for the nonlinear time trend in IMR (SI Appendix, Fig. S1). We kept only the SNPs with a missing call rate 0.01, minor allele frequency 0.01, and Hardy–Weinberg equilibrium test P value 1.0e-6 in GWAS. We also applied LD score regression-based genomic control where we inflated the SE of SNP effects using to conservatively control unadjusted confounding in association tests.
To estimate the effect sizes for the top loci in each birth year cohort (Fig. 4A), we used only samples born in the same year to run the IMR GWAS. Although we restricted birth year cohort with n > 5,000, we could not use BOLT-LMM due to its technical limitations. Instead, we ran the GWAS for each birth year cohort using Plink (54) with sex, genotyping array, and top 20 principal components as covariates.
Heritability and Genetic Correlation Estimation.
We used LD score regression (34) implemented in the LDSC software to estimate heritability of IMR, quantify heritability enrichment in 52 baseline functional annotations, and estimate genetic correlations of birth year IMR with 50 complex traits using GWAS summary statistics as input. Details of the 50 traits used in genetic correlation analysis are shown in Dataset S4.
Adjusting for Confounding due to Regional Characteristics.
To assess the potential confounding from long-run county differences, we performed additional GWAS adjusting for 1) socioeconomic status measures including household income (data field 738; n = 283,663) and educational years (data field 6138; n = 327,252) and 2) lagged IMR (n = 330,340). We coded household income following Hill et al. (55) into five ordered categories and modeled it as a fixed effect in the IMR GWAS. “Education years” was coded following Lee et al. (48) and used as a continuous covariate in the GWAS.
Quantifying and Removing Participation Bias.
We performed two secondary analyses to account for participation bias in GWAS. First, we conditioned the analysis on participation status of the optional mental health questionnaire (MHQ) in UKB. We categorized GWAS samples into three strata: MHQ = 0 (received the invitation but did not participate; n = 111,276), MHQ = 1 (received the invitation and participated; n = 107,364), or “NA” (did not receive the invitation due to unavailable email addresses; n = 111,700). Samples who did not receive invitations are enriched for older age, lower education (56). Within each MHQ stratum, we performed GWAS on birth year IMR using the same settings described above for the main GWAS.
We also used genomic structural equation modeling (GSEM) (57) to remove participation bias. Our model jointly regressed three GWAS of participation of optional questionnaires, that is, MHQ, food frequency questionnaire, and physical activity study, in UKB on a latent factor F representing participation behavior (SI Appendix, Fig. S12). The latent factor F and IMR were then regressed on each SNP to estimate participation-adjusted genetic association with birth year IMR. Since the coefficients and SEs in GSEM were on the scale of standardized phenotypes, we transformed GSEM results to the same scale of BOLT-LMM outcomes for comparison.
To guard against the participation bias due to migration behavior and undersampling in counties far from the assessment centers, we repeated the IMR GWAS using only the counties with sufficient samples. We tabulated the number of samples born in each of the 65 counties, and then performed the IMR GWAS using only the counties with at least 5,000 samples. This gave us n = 269,085 total samples from 17 counties. We also performed an additional IMR GWAS using only the samples whose region of birth is identical to the region of current address (n = 209,852). Results are shown in SI Appendix, Fig. S14.
IMR GWAS with Shuffled County ID.
As a sensitivity analysis, we ran the regional IMR GWAS with county ID shuffled (SI Appendix, Fig. S6). When we perform the shuffling analysis, a county has a vector of IMR values for different years. This whole vector is randomly assigned to a different county after shuffling. Then, we follow the same procedure from our primary analysis to assigned each UKB individual an IMR value based on the county and year combination. This way, two participants born in the same year and in the same county will still have identical phenotype values in GWAS. Finally, there are n = 327,100 samples in the GWAS. Since three new counties were formed in 1964, they do not have IMR records before 1964. To maintain maximum possible samples, we excluded these three new counties when shuffling.
SDS Data and Analysis.
SDS were computed using 3,195 individuals from the UK10K project (36). To match alleles between SDS and IMR GWAS, we first chose effect alleles in GWAS to always have positive associations with birth year IMR. Then, we obtained trait-SDS (tSDS) by transforming SDS such that tSDS = SDS if the derived allele in SDS is the effect allele in GWAS and tSDS = −SDS if the derived allele in SDS is the noneffect allele in GWAS. We set tSDS to “NA” otherwise. Following Berg et al. (49), we estimated Spearman correlations between tSDS and GWAS z scores and used a block jackknife approach to obtain the SEs and P values for the Spearman correlation. To remove spurious correlations caused by unadjusted confounding (e.g., population stratification), we also applied LD score regression (40) to compute genetic correlations between SDS and IMR GWAS associations.
To remove from IMR GWAS the population stratification captured by SDS, we adjusted the IMR GWAS effect sizes by regressing out SDS using the following steps:
-
1)
Temporarily hold out the SNPs with P value of <0.1 in the IMR GWAS.
-
2)
Using the remaining SNPs, regress the effect size estimates against the SDS to get the regression coefficient for SDS.
-
3)
Update the effect size estimates for all the SNPs: raw effect size minus SDS times its regression coefficient from step 2.
Then, estimate the genetic correlations with 50 complex traits using the updated effect sizes.
Data on Air Raids during World War II.
Detailed data on German air raids on the United Kingdom during World War II were obtained from ref. 58. We accessed the coordinates of bombing events and marked their locations on the map of UK using the “sf” R package with the maps and spatial data for the UK downloaded from the “GADM” (Database of Global Administrative Areas) website (59). We defined bombing density as the number of bombing events with casualty during 1940–1942 within a 10-km radius of each UKB participant’s place of birth.
Supplementary Material
Acknowledgments
We gratefully acknowledge use of the facilities of the Center for Demography of Health and Aging at the University of Wisconsin–Madison, funded by National Institute on Aging Center Grant P30 AG017266. We thank members of the Social Genomics Working Group at the University of Wisconsin for helpful comments. This research has been conducted using the UK Biobank Resource under Application 57284. This work is based on data provided through https://www.visionofbritain.org.uk/ and uses historical material which is the copyright of the Great Britain Historical GIS Project and the University of Portsmouth.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117312119/-/DCSupplemental.
Data Availability
GWAS summary statistics for birth year IMR are available at http://qlu-lab.org/data.html or at the GWAS catalog: http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90095001-GCST90096000/GCST90095047. Previously published data were used for this work (60).
References
- 1.Milot E., et al. , Evidence for evolution in response to natural selection in a contemporary human population. Proc. Natl. Acad. Sci. U.S.A. 108, 17040–17045 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byars S. G., Ewbank D., Govindaraju D. R., Stearns S. C., Colloquium papers: Natural selection in a contemporary human population. Proc. Natl. Acad. Sci. U.S.A. 107, 1787–1792 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee N., Shi J., García-Closas M., Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 17, 392–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtiol A., Tropf F. C., Mills M. C., When genes and environment disagree: Making sense of trends in recent human evolution. Proc. Natl. Acad. Sci. U.S.A. 113, 7693–7695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sella G., Barton N. H., Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu. Rev. Genomics Hum. Genet. 20, 461–493 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Beauchamp J. P., Genetic evidence for natural selection in humans in the contemporary United States. Proc. Natl. Acad. Sci. U.S.A. 113, 7774–7779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley D., et al. , Assortative mating and differential fertility by phenotype and genotype across the 20th century. Proc. Natl. Acad. Sci. U.S.A. 113, 6647–6652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong A., et al. , Selection against variants in the genome associated with educational attainment. Proc. Natl. Acad. Sci. U.S.A. 114, E727–E732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills M. C., et al. ; eQTLGen Consortium; BIOS Consortium; Human Reproductive Behaviour Consortium, Identification of 371 genetic variants for age at first sex and birth linked to externalising behaviour. Nat. Hum. Behav. 5, 1717–1730 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathieson I., et al. , Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanjak J. S., Sidorenko J., Robinson M. R., Thornton K. R., Visscher P. M., Evidence of directional and stabilizing selection in contemporary humans. Proc. Natl. Acad. Sci. U.S.A. 115, 151–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdellaoui A., et al. , Genetic correlates of social stratification in Great Britain. Nat. Hum. Behav. 3, 1332–1342 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Wilcox A. J., Fertility and Pregnancy: An Epidemiologic Perspective (Oxford University Press, New York, 2010). [Google Scholar]
- 14.Heligman L., Pollard J. H., The age pattern of mortality. J. Inst. Actuar. 107, 49–80 (1980). [Google Scholar]
- 15.Habicht J. P., Yarbrough C., Lechtig A., Klein R. E., Relationship of birthweight, maternal nutrition and infant mortality. Nutr. Rep. Int. 7, 533–546 (1973). [PubMed] [Google Scholar]
- 16.Williams E. J., Embleton N. D., Bythell M., Ward Platt M. P., Berrington J. E., The changing profile of infant mortality from bacterial, viral and fungal infection over two decades. Acta Paediatr. 102, 999–1004 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Wegman M. E., Infant mortality in the 20th century, dramatic but uneven progress. J. Nutr. 131, 401S–408S (2001). [DOI] [PubMed] [Google Scholar]
- 18.Vaupel J. W., Yashin A. I., Heterogeneity’s ruses: Some surprising effects of selection on population dynamics. Am. Stat. 39, 176–185 (1985). [PubMed] [Google Scholar]
- 19.Crimmins E. M., Finch C. E., Infection, inflammation, height, and longevity. Proc. Natl. Acad. Sci. U.S.A. 103, 498–503 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palloni A., Beltrán-Sánchez H., Discrete Barker frailty and warped mortality dynamics at older ages. Demography 54, 655–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruckner T. A., Catalano R., Selection in utero and population health: Theory and typology of research. SSM Popul. Health 5, 101–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobles J., Hamoudi A., Detecting the effects of early-life exposures: Why fecundity matters. Popul. Res. Policy Rev. 38, 783–809 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catalano R., Bruckner T., Smith K. R., Ambient temperature predicts sex ratios and male longevity. Proc. Natl. Acad. Sci. U.S.A. 105, 2244–2247 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James W. H., Grech V., A review of the established and suspected causes of variations in human sex ratio at birth. Early Hum. Dev. 109, 50–56 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Bozzoli C., Deaton A., Quintana-Domeque C., Adult height and childhood disease. Demography 46, 647–669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang C., Li Z., Narayan K. M., Williamson D. F., Martorell R., Bigger babies born to women survivors of the 1959–1961 Chinese famine: A puzzle due to survival selection? J. Dev. Orig. Health Dis. 1, 412–418 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Gørgens T., Meng X., Vaithianathan R., Stunting and selection effects of famine: A case study of the Great Chinese Famine. J. Dev. Econ. 97, 99–111 (2012). [Google Scholar]
- 28.Lührmann M., Wilson T., Long-Run Health and Mortality Effects of Exposure to Universal Health Care at Birth (Institute for Fiscal Studies, 2018). [Google Scholar]
- 29.Knox P. L., Convergence and divergence in regional patterns of infant mortality in the United Kingdom from 1949–51 to 1970–72. Soc. Sci. Med. D 15, 323–328 (1981). [DOI] [PubMed] [Google Scholar]
- 30.Lee C. H., Regional inequalities in infant mortality in Britain, 1861–1971: Patterns and hypotheses. Popul. Stud. (Camb.) 45, 55–65 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Gazeley I., Newell A., Reynolds K., Rufrancos H., How hungry were the poor in late 1930s Britain? Econ. Hist. Rev. 75, 80–110 (2022). [Google Scholar]
- 32.Boudreau F. G., Nutrition in war and peace. Milbank Mem. Fund Q. 25, 231–246 (1947). [PubMed] [Google Scholar]
- 33.Loh P.-R., et al. , Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan B. K., et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium, LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bersaglieri T., et al. , Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Field Y., et al. , Detection of human adaptation during the past 2000 years. Science 354, 760–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barreiro L. B., et al. , Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 5, e1000562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uciechowski P., et al. , Susceptibility to tuberculosis is associated with TLR1 polymorphisms resulting in a lack of TLR1 cell surface expression. J. Leukoc. Biol. 90, 377–388 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Wong S. H., et al. , Leprosy and the adaptation of human toll-like receptor 1. PLoS Pathog. 6, e1000979 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan B., et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., et al. , Estimating genetic nurture with summary statistics of multigenerational genome-wide association studies. Proc. Natl. Acad. Sci. U.S.A. 118, e2023184118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butie C., Matthes K. L., Hösli I., Floris J., Staub K., Impact of World War 1 on placenta weight, birth weight and other anthropometric parameters of neonatal health. Placenta 100, 150–158 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Stein A. D., Zybert P. A., van de Bor M., Lumey L. H., Intrauterine famine exposure and body proportions at birth: The Dutch Hunger Winter. Int. J. Epidemiol. 33, 831–836 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Saupe T., et al. , Ancient genomes reveal structural shifts after the arrival of Steppe-related ancestry in the Italian Peninsula. Curr. Biol. 31, 2576–2591.e12 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Kong A., et al. , The nature of nurture: Effects of parental genotypes. Science 359, 424–428 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Pirastu N., et al. ; FinnGen Study; 23andMe Research Team; iPSYCH Consortium, Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. 53, 663–671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyrrell J., et al. , Genetic predictors of participation in optional components of UK Biobank. Nat. Commun. 12, 886 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J. J., et al. ; 23andMe Research Team; COGENT (Cognitive Genomics Consortium); Social Science Genetic Association Consortium, Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berg J. J., et al. , Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sohail M., et al. , Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8, e39702 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao B., Yang C., Liu J., Zhou X., Accurate genetic and environmental covariance estimation with composite likelihood in genome-wide association studies. PLoS Genet. 17, e1009293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.University of Portsmouth, A vision of Britain through time. https://www.visionofbritain.org.uk. Accessed 12 July 2021.
- 53.Manichaikul A., et al. , Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hill W. D., et al. , Genome-wide analysis identifies molecular systems and 149 genetic loci associated with income. Nat. Commun. 10, 5741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams M. J., et al. , Factors associated with sharing e-mail information and mental health survey participation in large population cohorts. Int. J. Epidemiol. 49, 410–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grotzinger A. D., et al. , Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.War, State and Society, Bombing Britain. https://www.warstateandsociety.com/Bombing-Britain. Accessed 18 July 2021.
- 59.GADM, GADM data (version 3.6). https://gadm.org/download_country_v3.html. Accessed 25 May 2021.
- 60.Bycroft C., et al. , The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GWAS summary statistics for birth year IMR are available at http://qlu-lab.org/data.html or at the GWAS catalog: http://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/GCST90095001-GCST90096000/GCST90095047. Previously published data were used for this work (60).