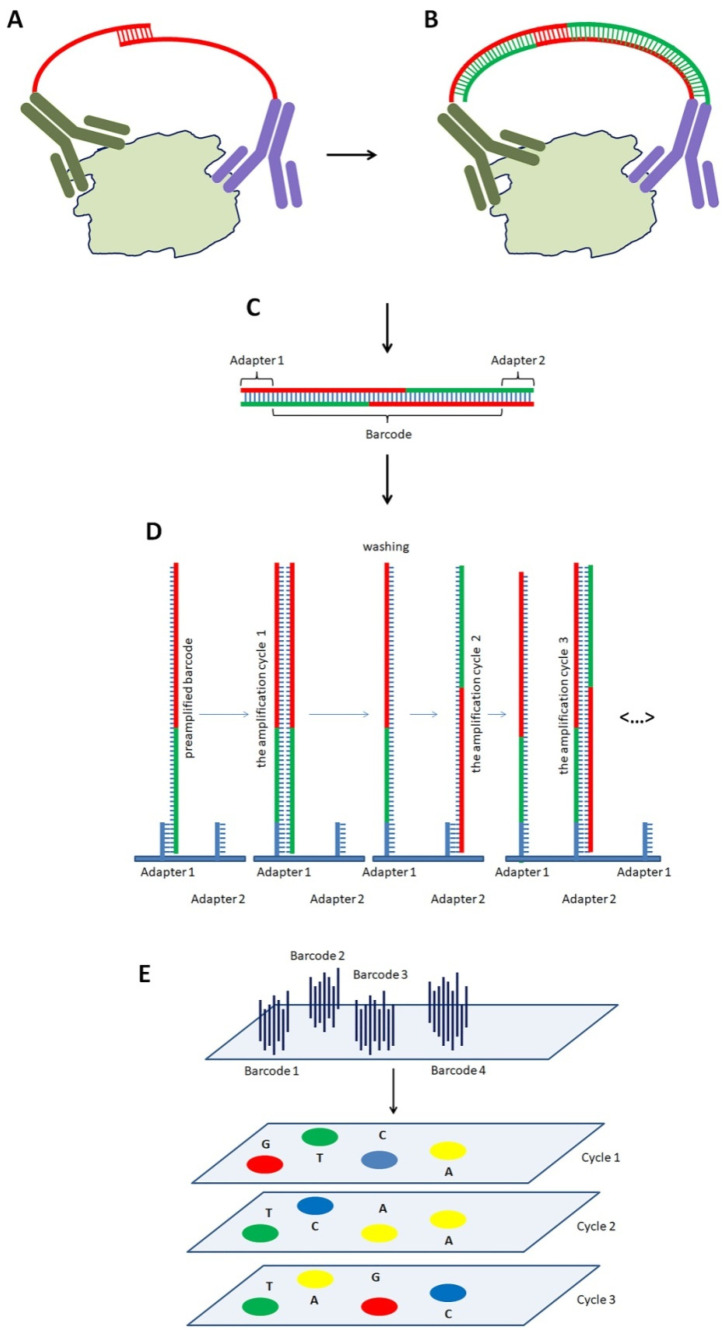
Figure 6.
The Proximity Extension Assay. The experiment includes several steps. (A). Immunoassay (Step 1). The pairs of specific antibodies conjugated with ssDNA oligonucleotides specifically interact with target proteins. As both ssDNA contain short complementary sequences, they interact and form a duplex in the middle. (B). Extension of ssDNA (Step 2). The duplex serves as a set of primers to DNA polymerase that extends ssDNA to dsDNA. (C). A cleavage of dsDNA (Step 3). Cutting dsDNA from the antibodies produces an oligonucleotide. Each oligonucleotide contains a unique sequence that serves as a barcode to identify the target protein. It also has two short adapter sequences at the ends. (D). Preparation for the sequencing of the barcodes (Step 4). The obtained double-stranded oligonucleotides are denatured and interact with immobilized probes complementary to the adapters. Then, DNA polymerase amplifies DNA using the probes as primers. (E). Clusterization of DNA and sequencing the barcodes (Step 5). As the concentration of DNA is relatively low, the amplified molecules form clusters. Each cluster originates from a single DNA molecule and represents a specific barcode. As the “clusters” have to be homogenous on their composition, the dsDNA is denatured to wash out the disconnected ssDNA. In addition, DNA attached to the slide through one of the adapters (e.g., adapter 2) becomes cut and removed. The following sequencing of the DNA identifies and quantifies the barcodes. The numbers of identified barcodes are proportional to the levels of the corresponding target proteins since the single interaction of specific antibodies to a target protein produces only one barcode.