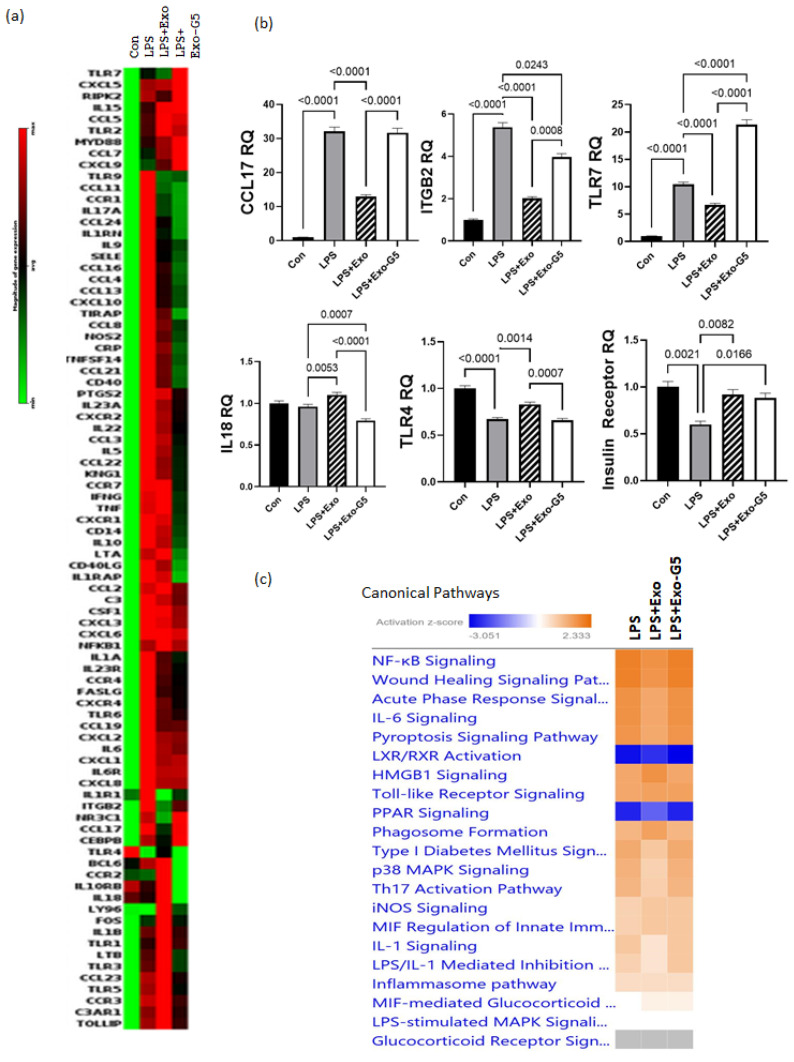
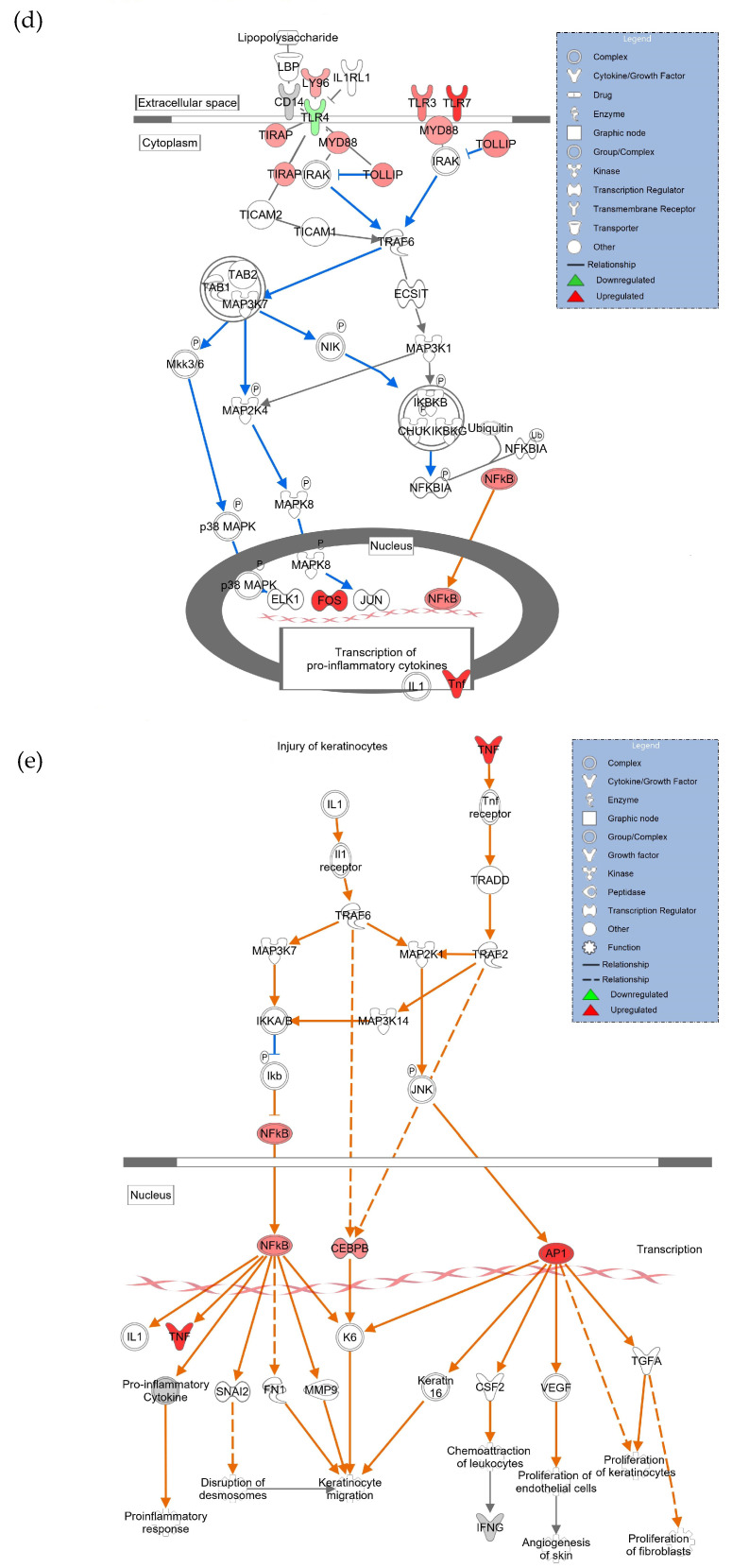
Figure 4.
HDF cells were treated with 5 ng/mL LPS for 6 h followed by hASC exosomes or exosomes depleted of GAS5 for 18 h. LPS was maintained in the media and cells were harvested after 4 days. RNA was isolated and PCR was run using the Human Inflammation Array (Qiagen, cat #PAHS-077Z). (a) Using GeneGlobe online data analysis software (Qiagen), a heatmap was generated from array data showing differentially expressed genes. (b) Genes identified from the heatmap showing patterns correlating to LPS, Exo, and Exo-G5 treatments were verified further by real-time qPCR. Relative quantification (RQ) was determined using a control sample as reference (n = 3). Statistical analysis was performed by one-way ANOVA and significant p-values (<0.05) are indicated on the graph. (c) ingenuity pathway analysis was performed on array data identifying canonical pathways influenced by GAS5 presence in exosomes. Changes in (d) the TLR pathway and (e) wound healing pathway were significantly altered in response to exosome and GAS5-depleted exosome treatment.