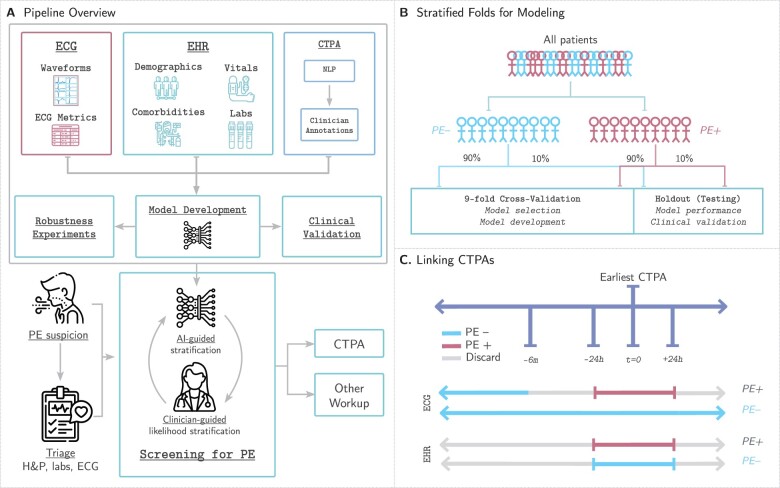
Figure 1.
Study design. (A) Our pipeline for creating models to detect pulmonary embolism consists of using three data modalities: electrocardiograms, clinical data [electronic health records (EHR)] including patient demographics, comorbidities, vital signs, and relevant labs, and computed tomography pulmonary angiograms that are labelled using a two-stage approach combining natural language processing pattern matching and manual clinician annotations. These data are linked together to develop, analyse, and benchmark models to predict pulmonary embolism. (B) We split our dataset for training, validation, and testing first by first identifying all unique patients (not unique computed tomography pulmonary angiogram or unique electrocardiogram) and separating them based on whether they have at least one PE-positive computed tomography pulmonary angiogram scan (PE+) or not (PE−). This stratum is further split into 90% for nine-fold cross-validation (89% for training, 11% for model selection and model development) and 10% for testing to assess model performance and benchmark against clinical scores. (C) Electrocardiograms are labelled as PE+ if they are recorded within 24 h of a PE+ computed tomography pulmonary angiogram. Electrocardiograms recorded 24 h after or between 6 months and 24 h before a positive computed tomography pulmonary angiogram are discarded. Electrocardiograms not meeting the above criteria for PE+ computed tomography pulmonary angiograms are labelled PE−. EHR data are retained if collected within 24 h of the computed tomography pulmonary angiogram and labelled equally with the computed tomography pulmonary angiogram finding.