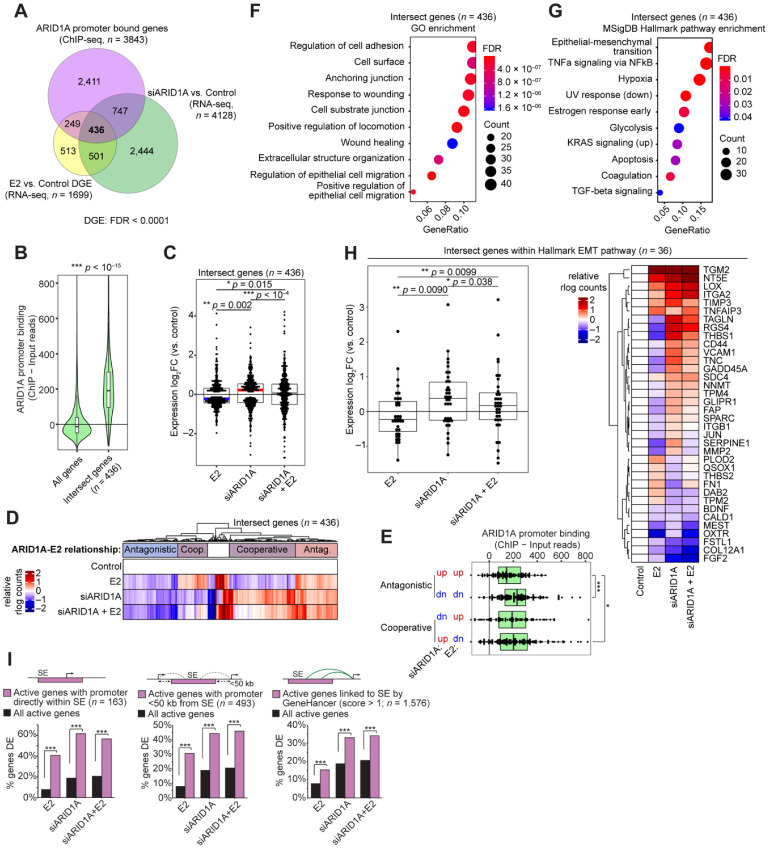
Figure 4.
ARID1A directly regulates estrogen-induced changes to cell identity. (A) Overlap between differentially expressed genes from E2 vs. Control (n = 1699, yellow) and siARID1A vs. Control (n = 4128, green) in 12Z-ESR1 cells and genes with ARID1A promoter binding by ChIP-seq in 12Z cells (n = 3843, pink) as previously described [24]. (B) Violin plot of ChIP signal for ARID1A binding at 436 intersect genes (as in A) compared to all genes. The unpaired, 2-tailed Wilcoxon was used for analysis. (C) Expression log2 fold-change values of 436 intersect genes following treatment with E2, siARID1A, or both E2 and siARID1A. The paired, 2-tailed Wilcoxon test was used for analysis. Box-and-whiskers plotted in the style of Tukey without outliers. (D) Clustering of 436 genes based on relative gene expression in each condition. Clusters have been labeled as antagonistic (red for upregulted or blue for downregulated) or cooperative (purple) in their regulation by E2 and ARID1A. (E) ARID1A binding among intersect genes segregated into 4 groups based on their direction of gene expression change following E2 or siARID1A treatment. (F) Pathway enrichment analysis of 436 intersect genes for Gene Ontology (GO) Biological Process. (G) Pathway enrichment analysis of 436 intersect genes for MSigDB Hallmark pathways. (H) Left, fold-change values of 36 Hallmark EMT genes found among the 436 intersect genes following treatment with E2, siARID1A, or both E2 and siARID1A. The statistic represented is from the paired, 2-tailed Wilcoxon test. Box-and-whiskers plotted in the style of Tukey without outliers. Right, relative expression of individual 36 EMT intersect genes as a clustered heatmap. (I) Enrichment of DE genes affected by E2 treatment, ARID1A loss, or both for genes with active promoters directly inside of super-enhancer (SE, pink) (n = 163) (left), promoters within 50 kb of a super-enhancer (n = 493) (center), or genes linked to super-enhancer by the GeneHancer database (n = 1576) (right) compared to enrichment for all active genes (black). Hypergeometric enrichment was used for analysis.