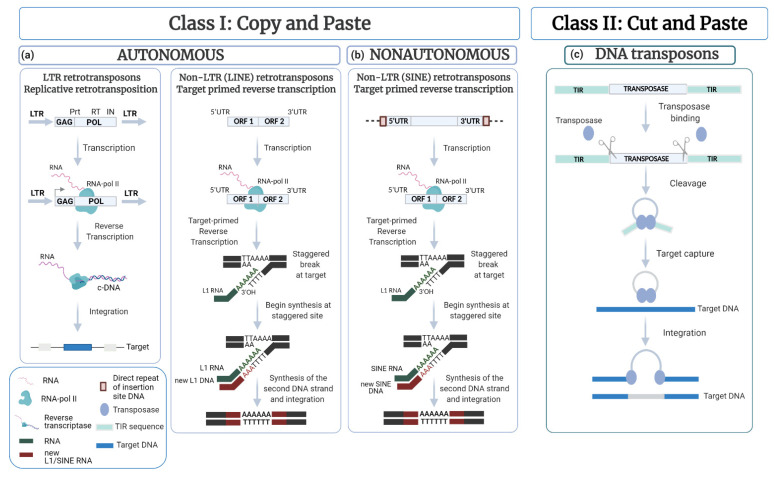
Figure 1.
Schematic representation of the different mechanisms of transposition. Generally, TEs can be distinguished in two major classes on the basis of their mechanism of transposition: active eukaryotic Class I (retrotransposons) and active eukaryotic Class II (DNA transposons). Additionally, TEs can be divided into autonomous and non-autonomous. (a) Class I (retrotransposons) require RNA transcription to be able to move to different genome locations. They encode for a reverse transcriptase enzyme that uses the transcript as a template to produce a cDNA sequence that reinserts randomly into a new genomic site. This is the so-called “copy and paste” mechanism. Autonomous Class I RNA transposons encode all proteins necessary for moving. They include long terminal repeats/endogenous retroviruses (LTR/ERV; e.g., the yeast Ty element) and non-LTR retrotransposons such as the long interspersed nuclear elements or LINEs (e.g., human L1). LTR-retrotransposons contain two long terminal repeats (LTRs, grey arrows) and genes encoding for functional proteins, such as Gag (group-specific antigen), Pol (reverse transcriptase), Int (integrase) and Prt (protease). The non-LTR retrotransposons also contain genes encoding for enzymes required for transposition but lack LTRs. Instead, they have two open reading frames flanked by a 5′ and a 3′ untranslated region (UTR). Generally, these TEs mobilize by a target-site primed reverse transcription (TPRT) mechanism. After the hydrolysis of one strand of DNA at a new insertion site, the 3′OH end of this strand is used to prime the reverse transcription of a new LINE cDNA by the reverse transcriptase encoded by the element. Subsequently, hydrolysis of the second DNA strand releasing a 3′OH end that primes replication of the second strand of the LINE cDNA. Finally, the integrase completes the insertion. (b) Non-autonomous retrotransposons rely on “true” (autonomous) retrotransposon activity for mobility. For example, SINE elements (like Alu) have an internal promoter for RNA polymerase III flanked by a 5′ and a 3′ UTR but lack genes encoding enzymes required for transposition. SINEs use the same TPRT mechanism to transpose, but they must borrow the necessary activity from LINE to insert. (c) Class II (DNA transposons) encode the protein transposase (TPase) flanked by terminal inverted repeats (TIRs). TPases are responsible for removing and inserting TEs in a new genomic location according to two different mechanisms. One is the so-called “cut and paste” or “non-replicative pathway” mechanism through which a TE is excised from its locus and reinserted at another site. The second is the “replicative pathway” in which a TE is copied, and the copy is relocated, leaving behind the original.