Fig 5. Upregulation of miRNA30b-30e in CD8+ T cells of HIV-infected individuals.
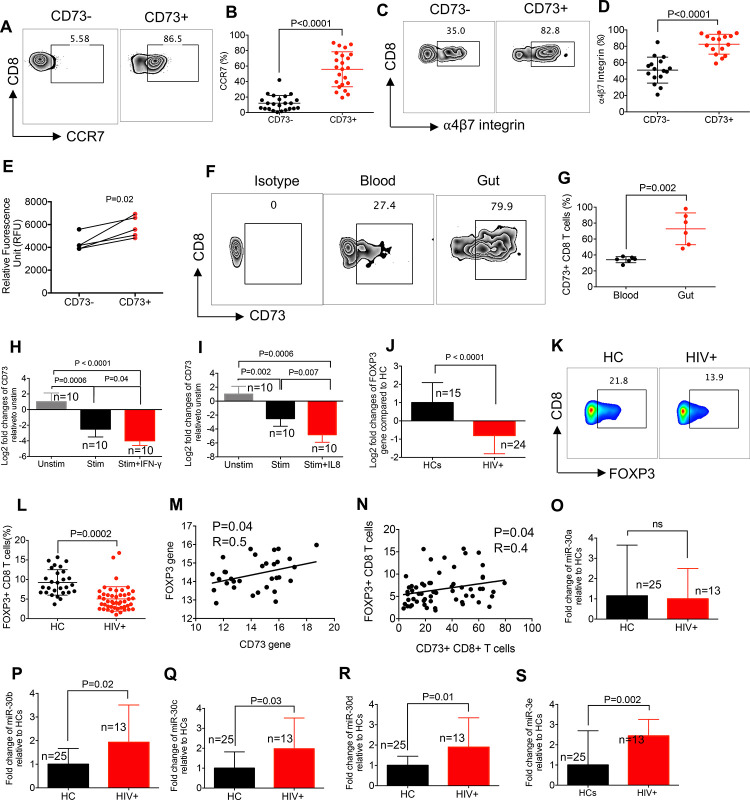
(A) Representative flow cytometry plots, and (B) cumulative data of CCR7 expression on CD73- versus CD73+ CD8+ T cells. (C) Representative flow cytometry plots, and (D) cumulative data of ⍺4β7 integrin expression on CD73- versus CD73+ CD8+ T cells. (E) Cumulative results of migratory ability of effector CD8+CD73- versus CD8+CD73+ T cells as measured by RFU. (F) Representative flow cytometry plots, and (G) cumulative data of the percentage of CD73 expressing CD8+ T cells in the peripheral blood and the gut tissue of mice. (H) Fold regulation of CD73 gene in isolated CD8+ T cells either unstimulated (unstim) or stimulated (stim) with anti-CD3/CD28 antibodies in the presence or absence of IFN-γ (100 ng/mL), and (I) IL-8 (100 ng/mL). (J) Fold regulation of FOXP3 gene in CD8+ T cells isolated from PMBCs of HIV-infected individuals relative to HCs quantified by qPCR. (K) Representative flow cytometry plots, and (L) cumulative data of FOXP3 expression in CD8+ T cells from HCs vs HIV-infected individuals. (M) Cumulative data of the correlation between the CD73 gene and the FOXP3 gene in CD8+ T cells of HIV-infected individuals and HCs. (N) Cumulative data of the correlation between the cell surface CD73 and FOXP3 expression in CD8+ T cells of HIV-infected individuals and HCs. (O) Fold change of miRNA-30a, (P) miRNA-30b, (Q) miRNA-30c, (R) miRNA-30d, and (S) miRNA30-e in CD8+ T cells of HIV-infected individuals relative to HCs quantified by qPCR. Each dot represents data from an individual human subject. Each dot represents data from an individual human subject. Statistical analysis determined by the Mann-Whitney U-test (B, D, E, G, J, L, and O-S), Kruskal–Wallis test (H and I), and by linear regression analysis (M and N) with significance indicated. ns: no significant. “n” reflects the number of samples.