Fig. 4.
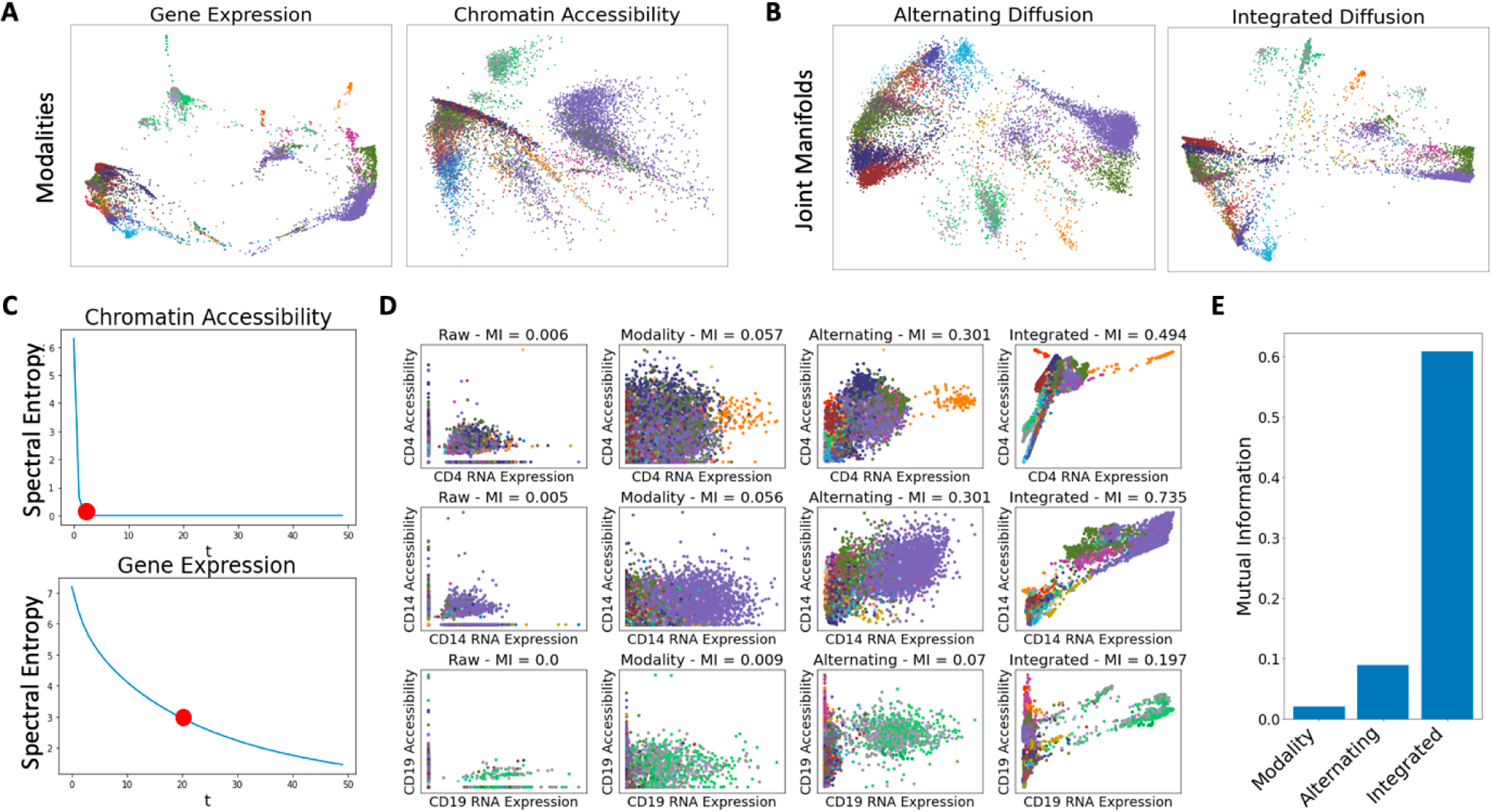
Application of integrated diffusion to multimodal single cell data. A) Visualization of gene expression and chromatin accessibility manifolds as well as B) alternating and integrated manifolds via PHATE. Points colored by annotated cell type. C) Visualization of spectral entropy of each modality. D) Mutual information between the expression of a gene and it’s accessibility across differing denoising strategies. E) Average mutual information for differing denoising strategies across all gene expression-gene accessibility pairs.
