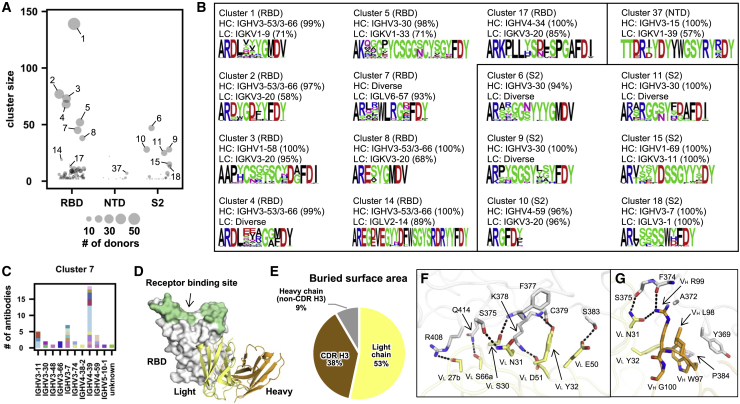
Figure 2.
SARS-CoV-2 S antibodies exhibit convergent CDR H3 sequences
(A) CDR H3 sequences from individual antibodies were clustered using a 80% sequence identity cutoff (see STAR Methods). The epitope of each CDR H3 cluster is classified based on that of its antibody members. Cluster size represents the number of antibodies within the cluster.
(B) The V gene usage and CDR H3 sequence are shown for each of the 16 CDR H3 clusters of interest. For each of the CDR H3 cluster of interest, the CDR H3 sequences are shown as a sequence logo, where the height of each letter represents the frequency of the corresponding amino-acid variant (single-letter amino-acid code) at the indicated position. The dominant germline V genes (>50% usage among all antibodies within a given CDR H3 cluster) are listed. Diverse: no germline V genes had >50% frequency among all antibodies within a given CDR H3 cluster. HC, heavy chain; LC, light chain. Clusters with the same domain specificity are grouped in the same box.
(C) IGHV usage in cluster 7 is shown. Different colors represent different donors. Unknown: IGHV information is not available.
(D) An overall view of SARS-CoV-2 RBD in complex with IGLV6-57 antibody S2A4 (PDB 7JVA) (Piccoli et al., 2020), which belongs to cluster 7, is shown. The RBD is in white with the receptor-binding site highlighted in green. The heavy and light chains of S2A4 are in orange and yellow, respectively.
(E) Percentages of the S2A4 epitope that are buried by the light chain, heavy chain (without CDR H3), and CDR H3 are shown as a pie chart. Buried surface area (BSA) was calculated by proteins, interfaces, structures, and assemblies (PISA) at the European Bioinformatics Institute (https://www.ebi.ac.uk/pdbe/prot_int/pistart.html) (Krissinel and Henrick, 2007).
(F and G) Detailed interactions between the (F) light and (G) heavy chains of S2A4 and SARS-CoV-2 RBD. Hydrogen bonds and salt bridges are represented by black dashed lines. The color coding is the same as (D). See also Figures S2–S4 and Tables S1 and S2.