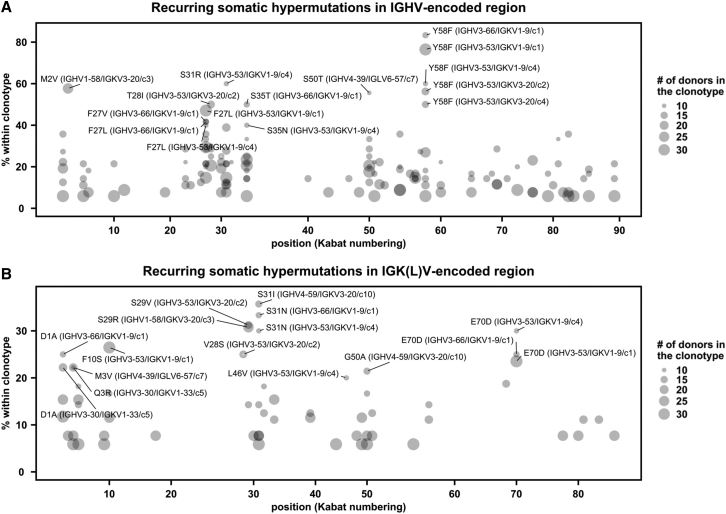
Figure 4.
SARS-CoV-2 S antibodies contain recurring somatic hypermutations (SHMs)
(A and B) For each public clonotype, if the exact same SHM emerged in at least two donors, such SHM is classified as a recurring SHM. Only those public clonotypes that can be observed in at least nine donors are shown. (A) Recurring SHMs in heavy-chain V genes. (B) Recurring SHMs in light-chain V genes. x axis represents the position on the V gene (Kabat numbering). y axis represents the percentage of donors who carry a given recurring SHM among those who carry the public clonotype of interest. For example, VL S29R emerged in 8 donors out of 26 donors that carry a public clonotype that is encoded by IGHV1-58/IGKV3-20. As a result, VL S29R (IGHV1-58/IGKV3-20) is 31% (8/26) within the corresponding clonotype. Of note, since each public clonotype is also defined by the similarity of CDR H3 (see STAR Methods), there could be multiple clonotypes with the same heavy- and light-chain V genes (e.g., IGHV3-53/IGKV1-9). The CDR H3 cluster ID for each clonotype is indicated with a prefix “c,” following the information of the V genes. For heavy chain, SHMs that emerged in at least 40% of the donors of the corresponding clonotype are labeled. For light chain, SHMs that emerged in at least 20% of the donors of the corresponding clonotype are labeled. See also Figure S5 and Table S1.