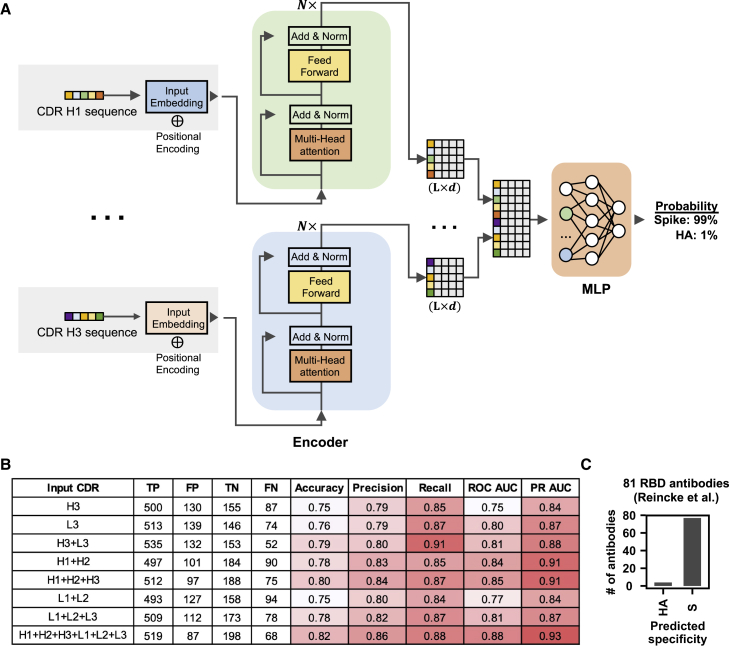
Figure 6.
Specificity of antibodies can be predicted by a sequence-based deep learning model
(A) A schematic overview of the deep learning model architecture.
(B) For evaluating model performance, S antibodies and HA antibodies were considered “positive” and “negative,” respectively. Model performance on the test set was compared when different input types were used. Of note, the test set has no overlap with the training set and the validation set, both of which were used to construct the deep learning model. True positive (TP) represents the number of S antibodies being correctly classified as S antibodies. False positive (FP) represents the number of HA antibodies being misclassified as S antibodies. True negative (TN) represents the number of HA antibodies being correctly classified as HA antibodies. False negative (FN) represents the number of S antibodies being misclassified as HA antibodies. See STAR Methods for the calculations of accuracy, precision, recall, ROC AUC, and PR AUC.
(C) The antigen specificity of 81 RBD antibodies from Reincke et al. (2022) were predicted by a deep learning model that was trained to distinguish between S antibodies and HA antibodies. See also Figure S6 and Table S3. The dataset for constructing and testing the deep-learning model, related to Figure 6, Table S4. Performance of the deep learning model with different inputs, related to Figure 6, Table S5. Prediction result of 81 antibodies to SARS-CoV-2 RBD that were elicited by Beta variant infection, related to Figure 6, Table S6. Prediction result of 691 HIV antibodies from GenBank, related to Figure 6.