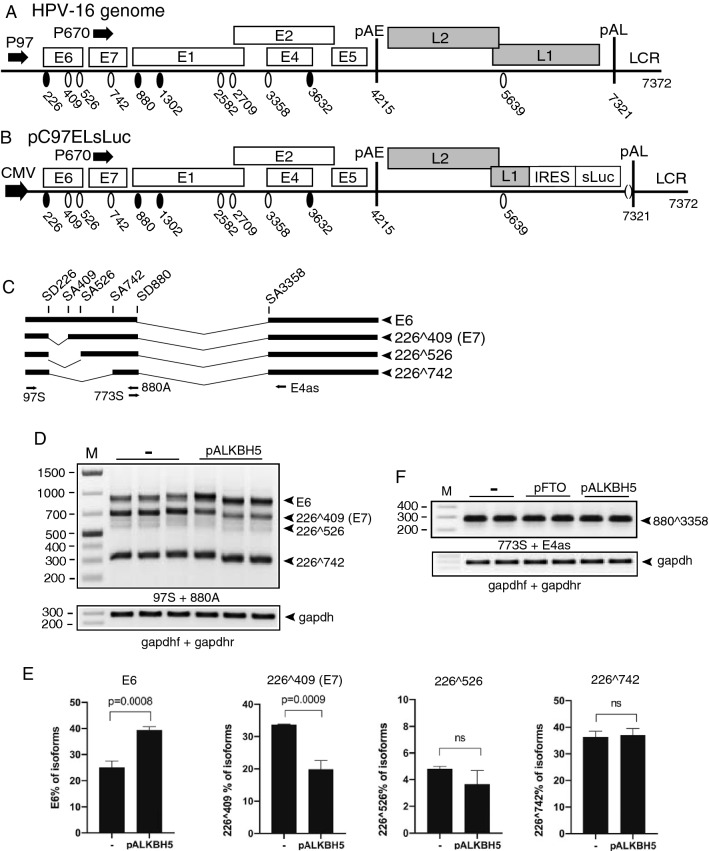
Fig. 1.
A Linearized HPV16 genome (numbers refer to the HPV16 reference strain GeneBank: K02718.1). Early and late genes are indicated. P97: HPV16 early promoter. P670: HPV16 late promoter. Black oval: splice donor. White oval: splice acceptor. pAE: HPV16 early polyadenylation site. pAL: HPV16 late polyadenylation site. LCR: HPV16 long control region. B HPV16 subgenomic plasmid pC97ELsLuc encodes all HPV16 genes. HPV16 early promoter P97 was replaced by human cytomegalovirus immediate early promoter (CMV). Secreted luciferase (sLuc) gene was integrated in the L1 gene following the poliovirus 2A internal ribosomal entry site (IRES) sequence. C Schematic structures of HPV16 early transcripts produced from pC97ELsLuc. Splicing at SD226 occurs independently of splicing at the downstream SD880 and generates splice variants in the E6- and E7-coding region. Arrows indicate HPV16 RT-PCR primers. D Effect of ALKBH5 on HPV16 E6/E7 mRNA splicing was monitored by RT-PCR with indicated primer pair on RNA extracted from triplicate transfections of pC97ELsLuc with empty pUC plasmid (−) or pALKBH5. E Densitometric quantitation of the results in E. The percentage of each indicated cDNA isoform of all cDNAs in a lane in the absence of ALKBH5 (−) or in the presence of ALKBH5 overexpression is displayed. F Effect of FTO or ALKBH5 on HPV16 E4 mRNA splicing was monitored by RT-PCR with indicated primer pair on RNA extracted from HeLa cells transfected with pC97ELsLuc and empty pUC plasmid (−), pFTO, or pALKBH5