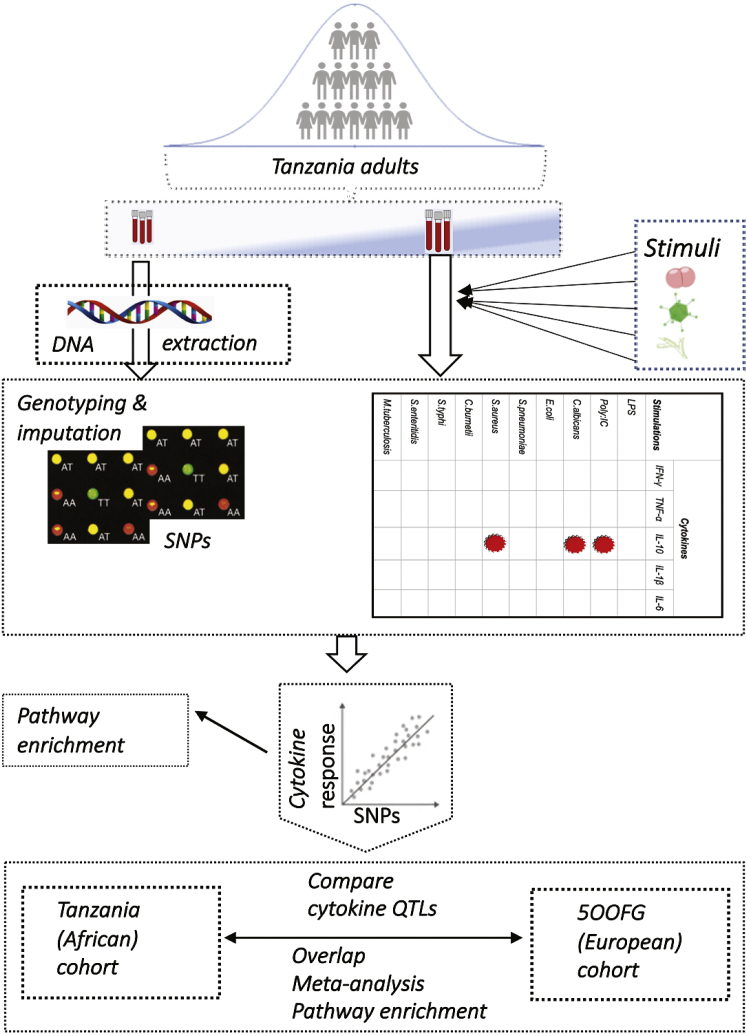
Figure 1.
Schematic diagram of study design and bioinformatics analyses
We utilized samples of a cohort of healthy Tanzanian adults. In vitro stimulation of their blood was performed with ten different stimuli, including bacterial, fungal, and TLR3 and TLR4 agonists followed by measuring five different cytokines in the supernatant. Three (indicated by red ovals) out of the resulting 50 cytokine-stimulation pairs were excluded from downstream analysis after quality control. Genotyping was performed using global screening arrays (GSAs) SNP chip. We performed genome-wide SNP cytokine quantitative trait loci (cQTL) mapping by correlating cytokine abundances and imputed genotype data and subsequently performed pathway enrichment analyses. Finally, we compared the results with another cohort of Western European ancestry individuals by using three different approaches: direct overlap of genetic variants, meta-analysis, and functional enrichment analysis.