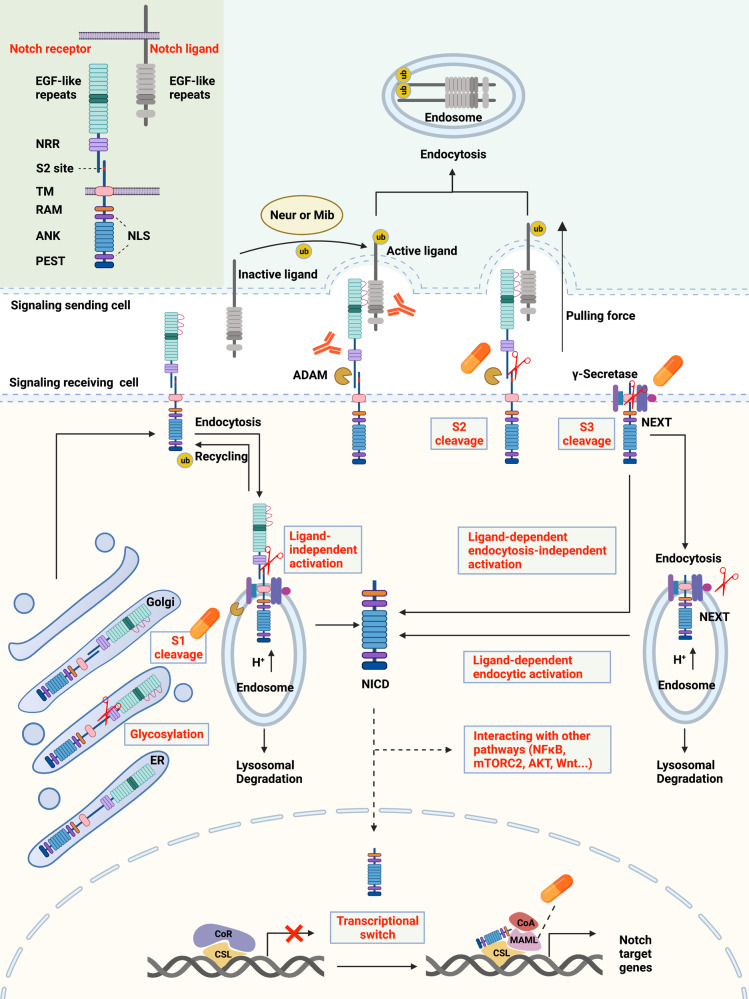
Fig. 2.
Overview of the NOTCH signaling pathway and therapeutic targets. In signal-receiving cells, NOTCH receptors are first generated in the ER and then trafficked to the Golgi apparatus. During trafficking, NOTCH receptors are glycosylated at the EGF-like repeat domain (red curves). Then, in the Golgi apparatus, NOTCH receptors are cleaved into heterodimers (S1 cleavage) and transported to the cell membrane. With the help of ubiquitin ligases, some of the NOTCH receptors on the cell membrane are endocytosed into endosomes. Endosomes contain an acidic environment with ADAMs and γ-secretase. The NOTCH receptors in endosomes can be recycled to the cell membrane, cleaved into NICD, or transported into lysosomes for degradation. In signal-sending cells, NOTCH ligands are distributed on the cell membrane and can bind to NOTCH receptors on signal-receiving cells. However, the ligands are inactive before ubiquitylation by Neur or Mib. After ubiquitylation, ligands can be endocytosed, thus producing a pulling force for the binding receptors. Without the pulling force, the S2 site (red marks) of NOTCH receptors is hidden by the NRR domain, and thus, the NOTCH receptors are resistant to cleavage by ADAMs. With the pulling force, the NRR domain is extended, therefore exposing the S2 site for cleavage. ADAMs and the pulling force are both necessary for S2 cleavage. After S2 cleavage, the remaining part of the NOTCH receptor is called NEXT. NEXT can be further cleaved on the cell membrane by γ-secretase or endocytosed into endosomes. In the former mode, NICD is released on the cell membrane. In the latter mode, NEXT can be cleaved into NICD or transported into lysosomes for degradation. In total, there are three approaches to generate NICD, classified as ligand-independent activation, ligand-dependent endocytosis-independent activation, and ligand-dependent endocytic activation. NICD can be translocated into the nucleus or remain in the cytoplasm to crosstalk with other signaling pathways, such as NFκB, mTORC2, AKT, and Wnt. The classical model proposes that, in the absence of NICD, CSL binds with corepressors to inhibit the transcription of target genes. Once NICD enters the nucleus, it can bind with CSL and recruit MAMLs, releasing corepressors, recruiting coactivators, and thus promoting the transcription of NOTCH target genes. There are two main approaches to inhibit NOTCH signaling for therapy. One is designing inhibitors of the key components of the pathways, including the enzymes that participate in S1 cleavage, ADAMs, γ-secretase, and MAML. The other one is producing antibody-drug conjugates against NOTCH receptors and ligands. The protein structures of NOTCH ligands and receptors are shown in the top left corner. NICD, NOTCH intracellular domain; ADAM, a disintegrin and metalloproteinase domain-containing protein; Neur, Neuralized; Mib, Mindbomb; NRR, negative regulatory region; NEXT, NOTCH extracellular truncation; CSL, CBF-1/suppressor of hairless/Lag1; MAMLs, Mastermind-like proteins; TM, transmembrane domain; RAM, RBPJ association module; ANK, ankyrin repeats; PEST, proline/glutamic acid/serine/threonine-rich motifs; NLS, nuclear localization sequence; CoR, corepressor; CoA, coactivator; ub, ubiquitin