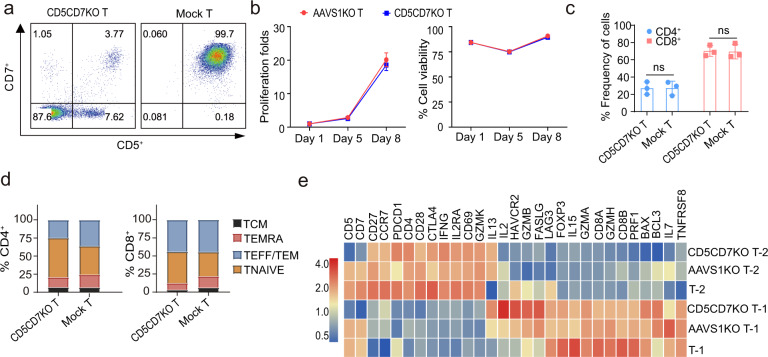
Fig. 1.
Disruption of CD5 and CD7 genes in T cells with CRISPR/Cas9 system. a Representative dot plots showing expression of CD5 and CD7 in CD5CD7KO T and mock T cells determined 3 days post electroporation with Cas9 protein, CD5 sgRNA-7, and CD7 sgRNA-85 using flow cytometry. b Proliferation and cell viability of CD5CD7 knockout T cells. Knockout of Adeno Associated Virus Integration Site 1(AAVS1), a safe harbor locus was used as a control. c The populations of CD4+/CD8+ T cells in CD5CD7KO T and mock T cells. The data represents mean ± SD for three donors. ns, no significance, two-way ANOVA. d The frequencies of effector and effector memory (TEFF/TEM; CCR7− CD45RA−), terminally differentiated effector memory (TEMRA; CCR7− CD45RA+), naïve (TNAIVE; CCR7+ CD45RA+), and central memory (TCM; CCR7+ CD45RA-) in CD5CD7KO T and mock T cells were measured using flow cytometry on days 7–10. The data represents the average from three donors. e Heatmap of selected genes related to T-cell activation, exhaustion, cytokine production, differentiation, and function-related genes