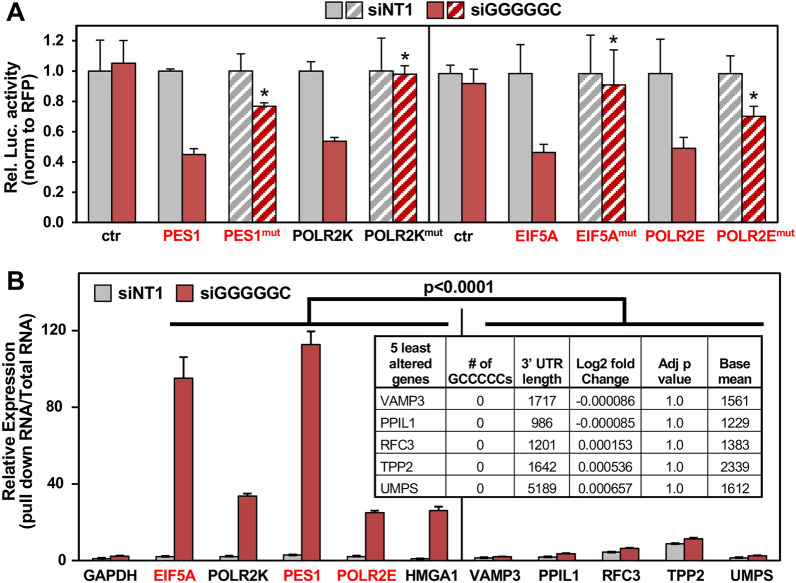
Figure 5.
Validation of GCCCCC seed matches located in the 3′ UTR of abundant genes being targeted by siGGGGGC. (A) Relative luciferase activity of control plasmid (pmiR-Target), wild-type and mutant 3′ UTR constructs of 4 of the genes in Fig. 4B in 293T cells. Data are shown as mean ± SD of three replicates. p-values (Student’s t test, *p < 0.05) represent comparison between change in relative luciferase of cells co-transfected with wt/mutant constructs (50 ng) and siGGGGGC (20 nM) normalized to cells transfected with siNT1. (B) Enrichment of siGGGGGC target mRNAs in biotin-streptavidin pulled down RNA normalized to input RNA in HeyA8 cells transfected with 50 nM Bi-siNT1 or Bi-siGGGGGC for 20 h by real-time PCR. Inset: Table showing 5 selected control genes (> 1000 normReads) expression of which did not change in RNA-seq data derived from HeyA8 cells transfected with 10 nM siGGGGGC vs. siNT1 for 24 h. Each bar represents ± SD of three replicates. Student’s t test was used to calculate p-value. Survival genes are shown in red.