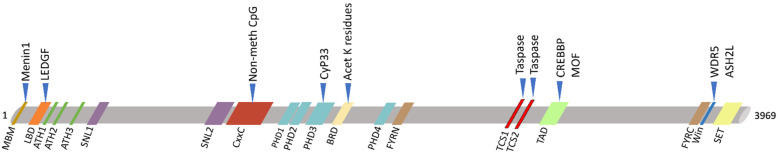
Figure 3.
Schematic view of KMT2A protein domains (below) and its main interactors (upper). KMT2A domains: MBM, high-affinity Menin-binding motif, residues 6–10; LBD, LEDGF-binding domain, residues 109–153; ATH1-2-3, AT-Hook1/2/3, residue 169–180, residues 217–227, residue 301–309; SNL1-2, nuclear-localization signal 1/2, residues 400–443, residues 1008–1106; CxxC, including: pre-CxxC region, residues 1149–1154, CxxC domain, residues 1147–1242, post-CxxC residues 1298–1337; PHD1-2-3-4, plant homology domain 1/2/3/4, residues 1431–1482, residues 1479–1533, residues 1566–1627, residues 1931–1978; BRD, bromodomain, residues 1703–1748; FYRN, FY-rich N-terminal domain, residues 2018–2074; TAD, transactivator domain, residues 2829–2883; FYRC, FY-rich C-terminal domain, residues 3666–3747; Win, WDR5 interaction motif, residues 3762–3773; SET, Su(Var)3-9 enhancer-of-zeste trithorax domain, residues 3829–2945. KMT2A has two sites for cutting by Taspase1: TCS1-2, taspase1 cleavage site 1/2, residue 2666–2670 and residues 2718–2722.