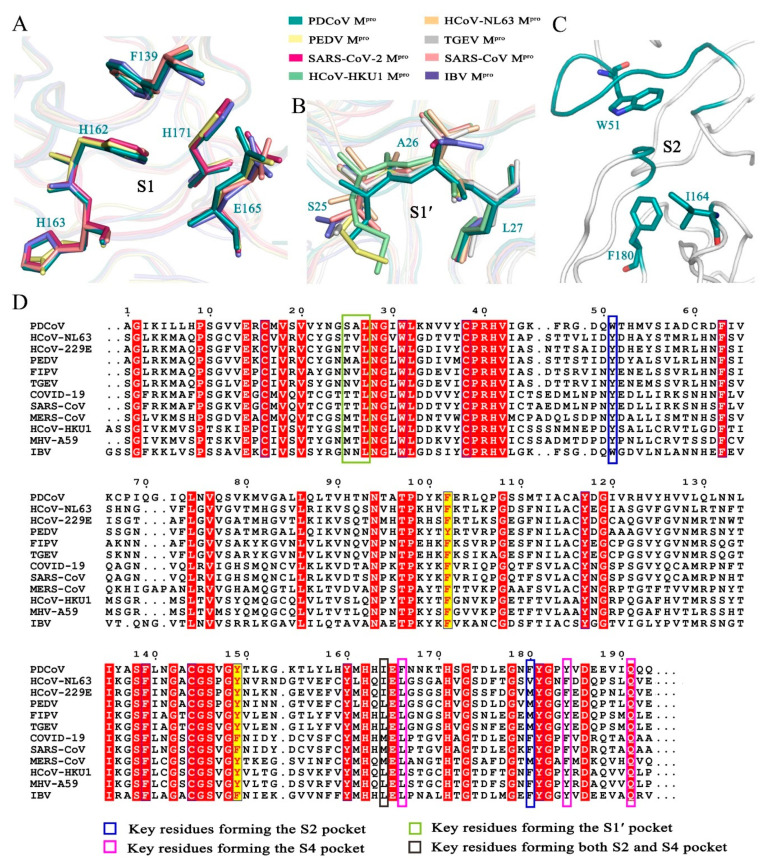
Figure 2.
Analysis of the substrate-binding pocket of PDCoV Mpro. (A) Superimposition of the key residues in the S1 binding pockets of PDCoV Mpro (deep teal), PEDV Mpro (pale yellow), SARS-CoV Mpro (salmon) and IBV Mpro (slate). (B) Superimposition of the key residues in the S1′ binding pocket of PDCoV Mpro (deep teal), HCoV-HKU1 Mpro (pale green), HCoV-NL63 Mpro (light orange), PEDV Mpro (pale yellow), SARS-CoV-2 Mpro (warm pink), SARS-CoV Mpro (salmon), TGEV Mpro (silver) and IBV Mpro (slate). (C) Hydrophobic residues in the S2 binding pocket. (D) Sequence alignment of the Mpro domains I and II from four different genera. PDCoV from Deltacoronavirus; HCoV-NL63, PEDV, FIPV, TGEV from Alphacoronavirus; SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, MHV-A59 from Betacoronavirus; IBV from Gammacoronavirus. A sequence alignment was generated using the program ClustalW and drawn by ESPript3. White letters with red backgrounds show identical residues, and red letters with yellow backgrounds show conservative variation.