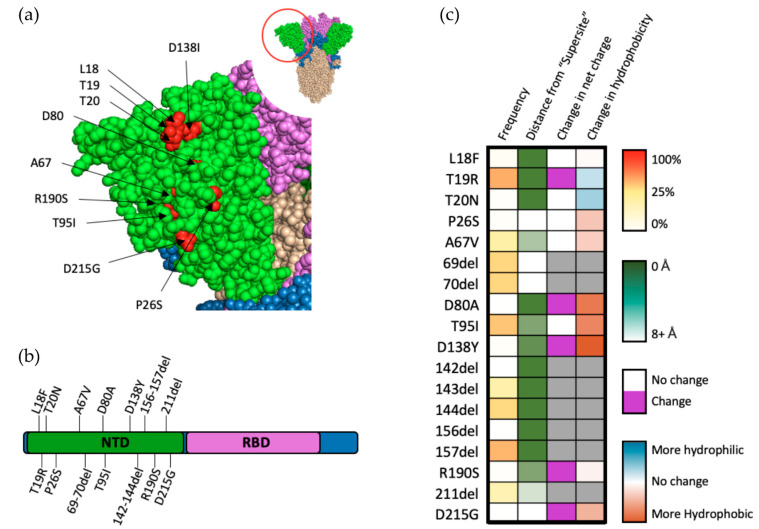
Figure 4.
Mutations within the NTD of SARS-CoV-2 spike protein. (a) Structural representation of substitutions (red) within the NTD (green) of SARS-CoV-2 spike protein. (b) Visual representation of mutations within the NTD of spike protein. (c) Frequency, residue distance from NTD “supersite”, modification in charge at physiological pH, and change in hydrophobicity at pH 7. Residue distance was calculated in PyMOL on the protein three-dimension structure (PDB-6ZGG), by measuring the distance between the nearest atom of “supersite” amino acids identified by Mccalum et al., 2021 [32] and the nearest atom of amino acids of interest.