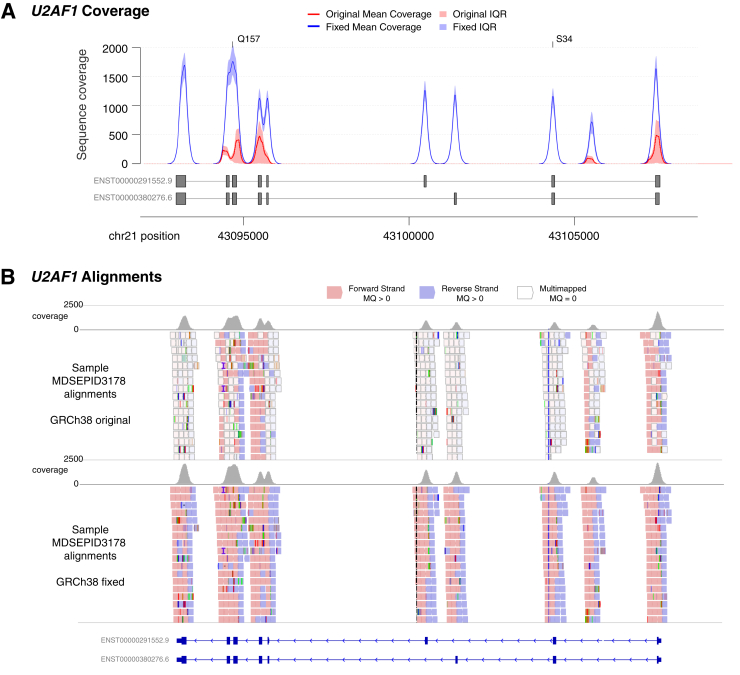
Figure 1.
Alignment issues across the U2AF1 locus. A: Sequence coverage of reads with mapping quality (MQ) >0 across the U2AF1 gene for 120 bone marrow samples sequenced after capture with a custom reagent. The mean coverage for realignments to GRCh38 are shown in red, whereas the mean coverage for alignments to the authors’ modified GRCh38 reference is shown in blue. Shading indicates the interquartile range (IQR) for each. Exons from the two primary protein-coding isoforms are shown below, and the locations of hotspot mutations at amino acids S34 and Q157 are indicated. B: An Integrated Genomics Viewer (IGV) view of sequence reads, with alignments to GRCh38 at top and alignments to the modified reference at bottom. Grey bars at top show overall coverage. Reads in white indicate multimapped reads, with mapping qualities of zero, whereas red and blue reads have higher quality alignments.