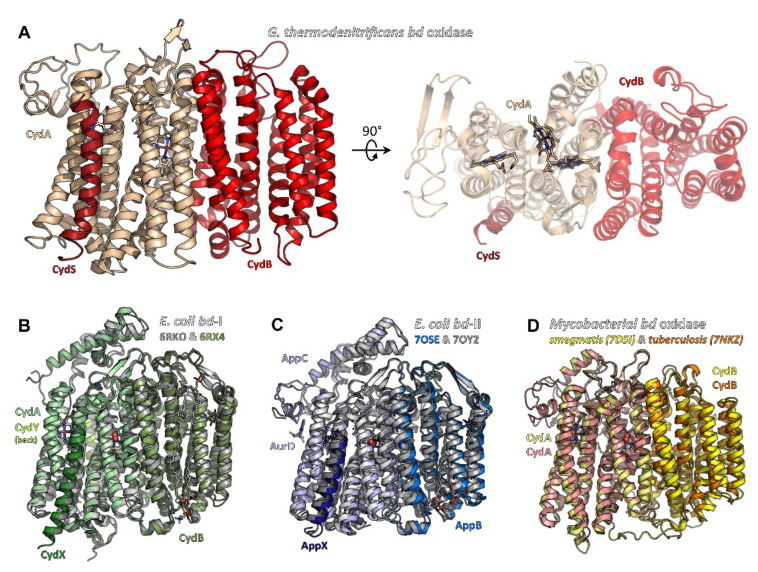
Figure 1.
Structures of bacterial cytochrome bd oxidases. (A) bd oxidase of G. thermodenitrificans (pdb ID 5DOQ) is composed of subunits CydA (light beige), CydB (red), and CydS (dark red). The heme groups are located in subunit CydA. (B) E. coli bd-I (pdb ID 6RKO in grey tones, pdb ID 6RXO in green colours) comprises four subunits, termed CydA, CydB, CydX, and CydY. While CydA, CydB, and CydX have homologues in G. thermodenitrificans, CydY is exclusive for E. coli bd-I. (C) bd-II oxidase from E. coli (pdb ID 7OSE in blue colours, pdb ID 7OY2 in grey tones) is built by subunits AppC (homologue to CydA), AppB (homologue to CydB), and AppX (homologous to CydS/X). 7OSE has been solved with the inhibitor aurachin D (AurD) bound to the Q-loop. (D) The mycobacterial bd oxidase (M. smegmatis, pdb ID 7D5I, in yellow colours, M. tuberculosis, pdb ID 7NKZ, in orange and salmon) consists of only two subunits, CydA and CydB.