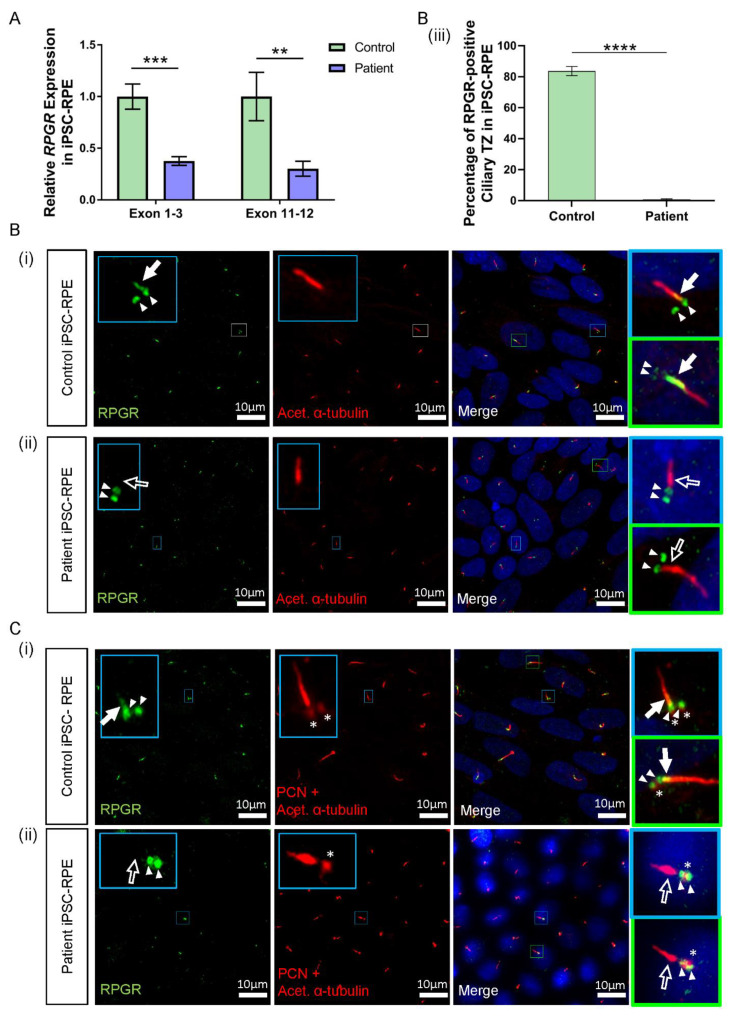
Figure 4.
RNA and protein studies in RPGR c.1415 − 9A>G variant iPSC-RPE cells. (A) The expression of RPGR determined by RT-qPCR was decreased in patient iPSC-RPE compared to control cells at exons 1–3 and exons 11–12 (unpaired t-test, ** p < 0.01, *** p < 0.001; SEM error bars from n = 4 independent experiments per line). Expression levels are relative to both HPRT and POLR2A housekeeper genes. (Controls = Controls 1 and 2; Patient = Clones 1 and 2). (B) (i) Control iPSC-RPE cells were stained with acetylated α-tubulin (red), which stained the primary cilium. Staining with RPGR (green) showed the presence of RPGR in the TZ region of the cilium (white arrow), with this region showing as yellow on the merged image (white arrow). Adjacent RPGR punctate staining was also present (paired white arrowheads). (Control = Control 1). (ii) Patient iPSC-RPE cells demonstrated a striking lack of RPGR staining in the cilium TZ region (white arrow outline), with residual adjacent punctate staining (paired white arrowheads). (Patient = Patient Clone 1). These findings are also demonstrated in (C) (i) and (ii). (iii) Quantification of the RPGR presence at the ciliary TZ confirmed decreased levels in patient cells compared with control lines. Values are expressed as a percentage of the total number of cilia present (unpaired t-test, **** p < 0.0001, SEM from n = 4 independent experiments per line, n = 159 control cilia counted, n = 219 patient cilia counted). (Control = Controls 1 and 2; Patient = Clones 1 and 2). (C) (i) In Control iPSC-RPE cells, localisation of the RPGR punctate staining (paired white arrowheads) was determined to be at the basal bodies by co-staining with PCN (red, white asterisks). (Control = Control 2). (ii) Patient iPSC-RPE cells also showed a co-localisation of RPGR staining with the PCN staining. (Patient = Clone 2).